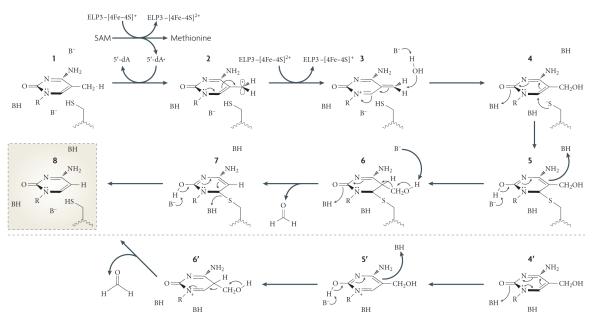
Figure 6. Proposed mechanism for elP3-mediated DNa demethylation.
Mammalian elongator complex protein 3 (ELP3) contains an Fe-S radical S-adenosylmethionine (SAM) domain that is important for active DNA demethylation of the zygotic paternal genome. If ELP3 is indeed a functional radical SAM protein, it may directly carry out DNA demethylation through the following mechanism. First, ELP3 uses SAM to generate a 5′-deoxyadenosyl radical, which could extract a hydrogen atom from the 5-methyl group of 5-methylcytosine (5meC; 1) to form a 5meC radical (2). After an electron is donated back to the Fe -S to create the third intermediate (3), a water molecule would promote the formation of 5-hydroxymethylcytosine (5hmC) (4). A nucleophilic attack at carbon 6 can result in the carbon-carbon bond breaking to release formaldehyde (5-7). In the absence of an external nucleophile, an alternative pathway (4′-6′) that leads to the release of formaldehyde can also take place. Finally, an elimination step would produce an end product of C (8).