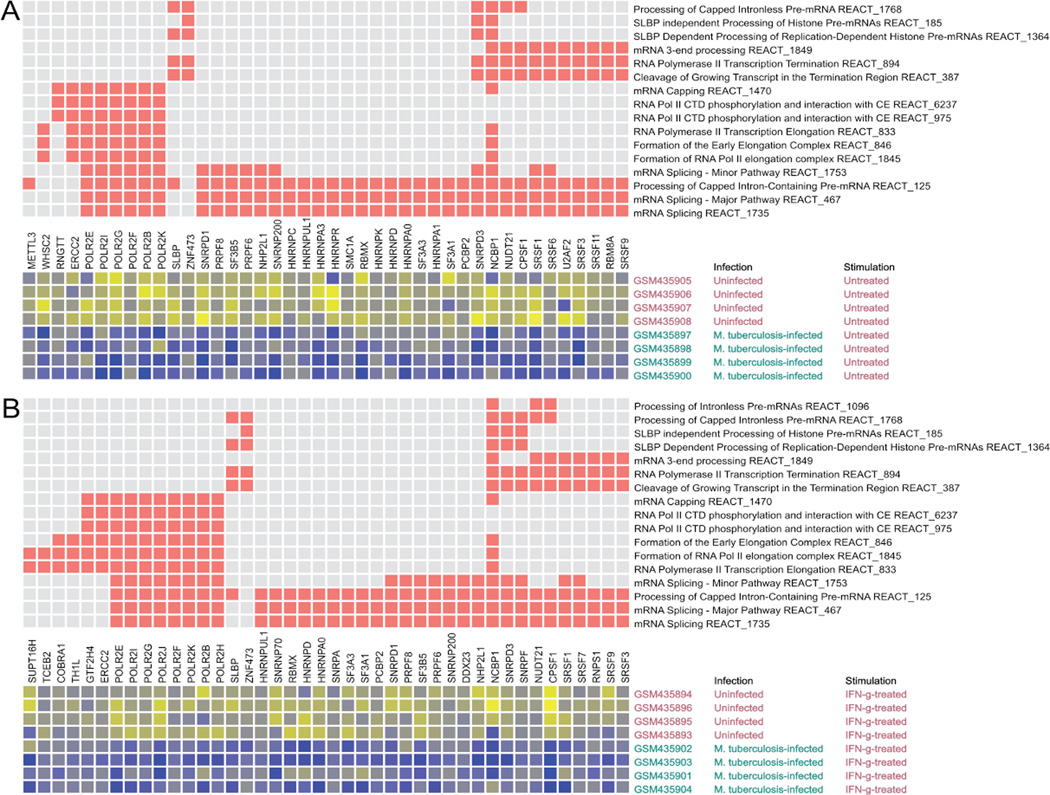
Figure 6.
Post-initiation mRNA biogenesis Reactome pathways and genes are down-regulated by M. tuberculosis infection of THP-1 cells. Eighteen pathways were tested. The pathways shown are significantly different for the indicated comparisons (A and B) based on a false discovery rate (FDR) criterion of 0.05 for the CERNO test results. The pathway tests were based on the rank order of differential expression for all genes in a pathway (among all transcript measurements of all named genes targeted by the gene expression platform). The top matrix depicts the membership of genes (columns) in pathways (rows) with red squares. The rows and columns of the top matrix are sorted to bring together similar pathway membership patterns. Genes shown are members of one or more pathways that exhibited differential expression for the indicated comparison (one-sided Student's T-test to assess down-regulation, p < 0.05 unless otherwise noted). The lower matrix is a heatmap of expression for each of the significantly regulated genes in each of the samples that were compared. The heatmap shows gradations from higher to lower expression as yellow to blue. The decrease in expression (blue) is evident for infected cells. Gene symbols are shown with the NCBI Gene IDs. A) 16 pathways were down-regulated when comparing infected THP-1 cells to uninfected cells. B) 17 pathways were down-regulated when comparing THP-1 cells infected with M. tuberculosis and stimulated with IFNγ to uninfected, IFN-γ-stimulated THP-1 cells.