Figure 2. Detection sensitivity of the magneto-DNA system.
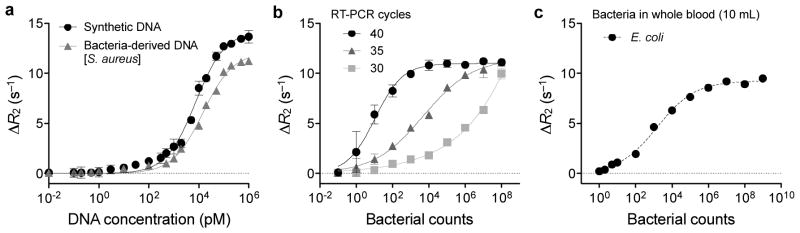
a, Serial dilutions of synthetic DNA or bacteria-derived DNA were used as detection targets. Bacteria-derived DNA molecules were obtained via asymmetric RT-PCR of S. aureus 16S rRNA (35 cycles); synthetic DNA had the same sequence as bacteria-derived DNA. The detection limit was ~ 0.5 pM. b, Bacterial detection by the magneto-DNA assay. Samples with varying numbers of S. aureus were used. Total RNA was extracted and target sequences were amplified by 30, 35, and 40 cycles of RT-PCR. Amplified target DNA were detected using the probe set in a. The observed detection limit was a single bacterium, and the dynamic range of detection could be controlled by changing the PCR cycle number. c, Bacterial detection in blood. Serial dilutions of E. coli were spiked into human blood and processed by first lysing the red blood cells and then extracting the RNA using the same procedure as described above. All experiments were done in triplicate. All ΔR2 values were obtained by subtracting the relaxation rate values of the hybridised probe complexes in the presence of target DNA (R2,target), with the relaxation rate values of the beads alone (R2,control). Data are expressed as mean ± SD.
