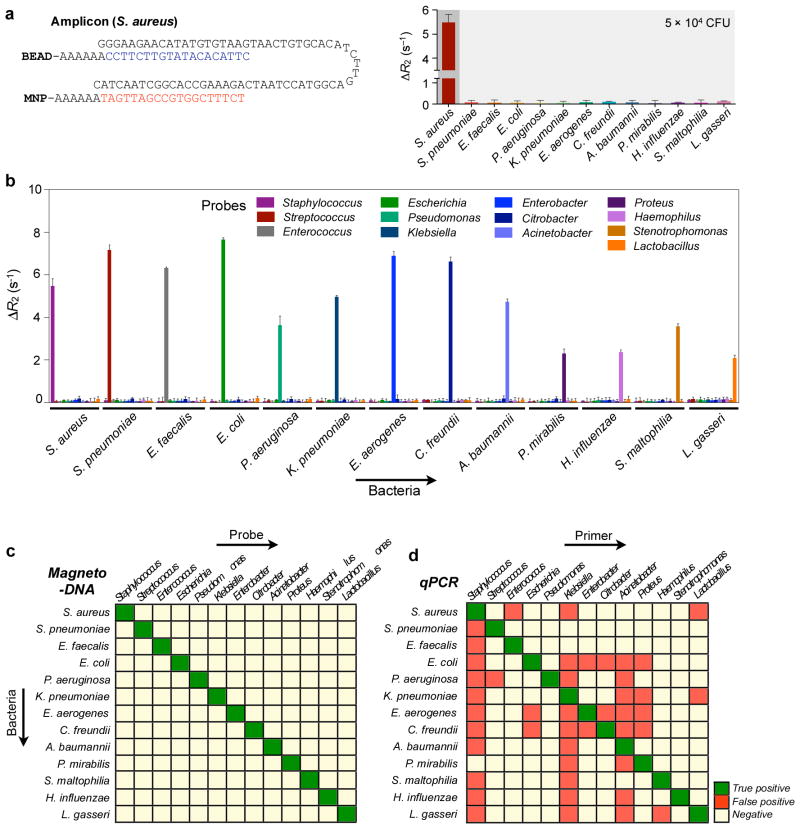
Figure 4. Differential detection using the magneto-DNA system.
Probes targeting hypervariable regions of bacterial 16S rRNA sequences were used to specifically detect various bacterial types. RNA was extracted from bacterial cultures, amplified by asymmetric RT-PCR (35 cycles) using specific primers for each species, and detected using the corresponding probe conjugates. a, Probes specific for Staphylococcus were used for detecting S. aureus (DNA amount equivalent to 50,000 CFU). Target DNA from other bacterial species were added as controls to test off-target binding of the probes. b, Relaxation rates for differential detection of various bacterial types. Note the high specific signals and low background noise against other bacteria. Data are expressed as mean ± SD. All samples for the assay were prepared in triplicate. c,d, Heat maps comparing the specificity of the magneto-DNA assay with that of qPCR. Specificities in c were based on ΔR2 values from the magneto-DNA assay shown in b. Specificities in d are relative target amounts obtained from qPCR in Figure S5. Significant signals were marked as positive: positive signals for specific target bacteria were regarded as ‘true-positive’, while positive signals from non-targeted samples were classed as ‘false-positive’.