Figure 9.
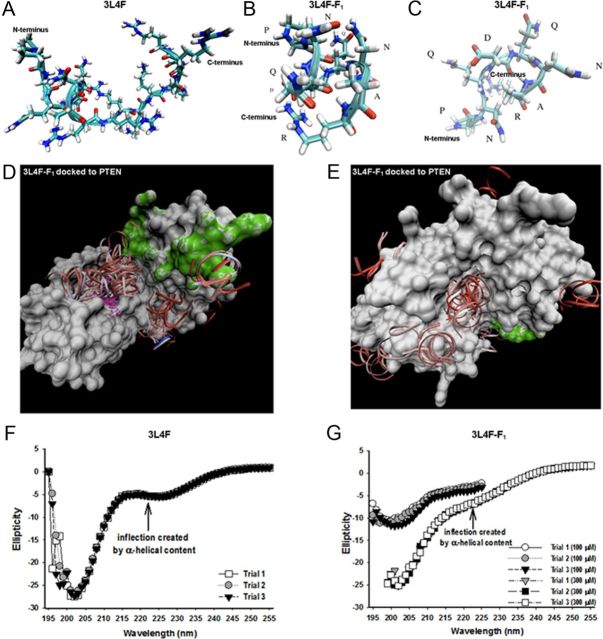
Molecular modeling of 3L4F and 3L4F-F1 peptides against PTEN predict structural elements important for PTEN recognition and inhibition. A, B, The predicted conformation as derived from REMD simulations of the 3L4F peptide (A) displays α-helical content at the N terminus, which is conserved in the 3L4F-F1 peptide (B) conformations. C, The 3L4F-F1 peptide REMD simulations also predicted β-turn structure. D, GRAMM dockings of the most populated α-helical conformation clustered in the central groove between the phosphatase active site and C2 membrane-targeting structural domains. The phosphatase active site of PTEN is denoted in purple, and the C2 membrane-targeting structural domain of PTEN is denoted in green. E, GRAMM dockings of a less-populated cluster site of α-helical conformations was found on the immediately opposite side of the central groove. CD detects the presence of some α-helical content (inflection point at 222 nm) in the 3L4F (300 μm) (F) and 3L4F-F1 peptide (100 and 300 μm) (G). Each peptide was subjected to three trial runs of CD.