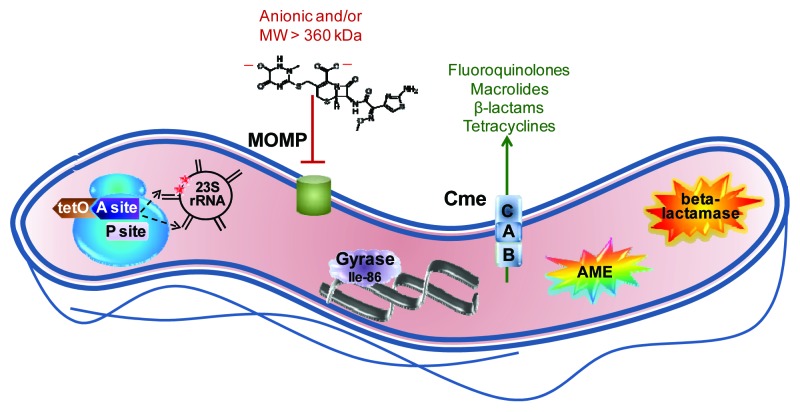
Figure 1. Summary of major antibiotic resistance mechanisms in Campylobacter. The ribosome, shown in blue at the left, is the site of two major resistance mechanisms. Binding of the TetO protein (shown in brown) to the A site (shown in dark purple) prevents tetracycline from occupying that site but stills allows access of the aminoacyl tRNA so that protein synthesis continues. Point mutations in 23S rRNA in the domain V region (shown in black) at position 2,075 principally and less often at position 2,074 (indicated by red stars) decrease the binding affinity for macrolides and lead to resistance. The major outer membrane protein (MOMP, shown in green), limits the entry of most antibiotics that are negatively charged or with a molecular weight larger than 360 kDa; the structure of the 552 kDa, dianionic antibiotic ceftriaxone is shown as an example. The Thr-86-Ile substitution in DNA gyrase (shown in light purple), is the main means of fluoroquinolone resistance, and this single mutation also confers high level resistance to this antibiotic class. The multi-drug efflux pump CmeABC (shown as stacked blue squares) contributes to resistance against fluoroquinolones, macrolides, β-lactams and tetracyclines, and works synergistically with other resistance mechanisms, often leading to high-level resistance. Aminoglycoside-modifying enzymes (AME; shown as the multi-colored star burst), principally of the aminoglycoside phosphotransferase family, are the main means of aminoglycoside resistance. Finally, β-lactamases (shown as the orange star burst) of the penicillinase type as well as the Ambler class D OXA-61 contribute to β-lactam resistance.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.