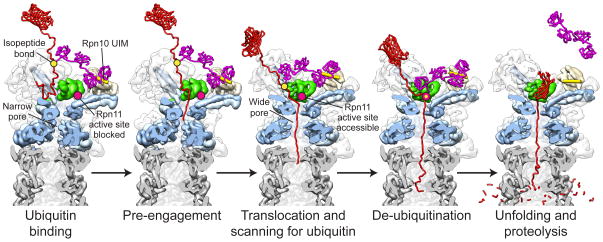
Figure 8. Structure-based model for substrate engagement and degradation by the 26S proteasome.
Cutaway side view of the proteasome reconstructions in the substrate-free and engaged conformations. In the first step, substrate (red) is tethered through its ubiquitin chain (purple) to the UIM of Rpn10 (yellow cylinder). In this pre-engaged state, the flexible substrate tail can enter the accessible N-ring pore and contact the uppermost subunits of the AAA+ domain spiral staircase. Upon substrate engagement, the Rpts become rearranged into a new spiral staircase with a widened central pore that is aligned with the N-ring and subjacent peptidase (grey). Concomitantly, Rpn11 (green) shifts to a central location directly above the N-ring pore, exposing its active site (pink dot) for ubiquitin scanning along the translocating polypeptide. All ubiquitin modifications are removed as their isopeptide attachment site (yellow dot) passes by Rpn11, facilitating fast translocation, unfolding, and degradation of the substrate.