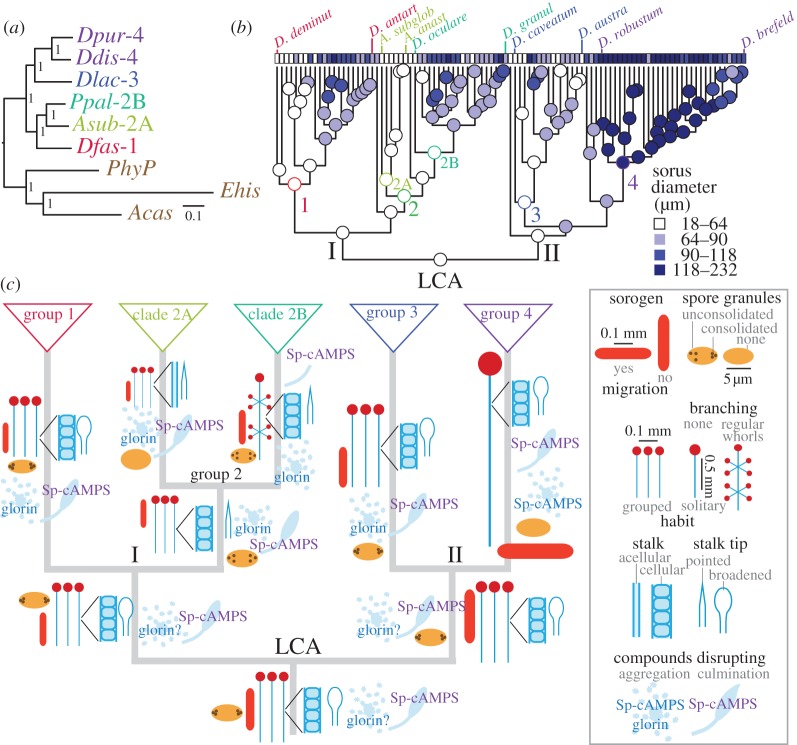
Figure 3.
Phylogeny correction and ancestral state reconstruction. (a) Genome-based core phylogeny. The sequences of 32 orthologous proteins in six group- or clade-representative Dictyostelid taxa (Ddis/Dpur—group 4, Dlac—group 3, Ppal—clade 2B, Asub—clade 2A, Dfas—group 1) and three outgroup taxa Ehis, Acas and PhyP were retrieved from genome sequencing projects, aligned and concatenated. The full alignment and subsets thereof (see the electronic supplementary material, figure S4) were subjected to Bayesian inference for phylogeny reconstruction [17]. Bayesian posterior probabilities of nodes are indicated. Scale bar: number of substitutions per site. (b) Character evolution. All measured characters were combined with the newly inferred phylogeny for all Dictyostelia (see the electronic supplementary material, figure S5) and subjected to inference of character history and ancestral state reconstruction using maximum-likelihood-based methods. The analysis of quantitative characters is listed in Excel file ‘Trait_Analysis’, sheet 6 and of categorical characters in the electronic supplementary material, figure S6a–p. For graphical representation of the evolutionary history of the character ‘sorus diameter’, the range of calculated values was subdivided into four intervals, which, represented by shades of blue, were plotted onto the phylogeny. (c) Ancestral states at major nodes. For quantitative characters, the state values at nodes that connect major branches (highlighted in colour in ‘Trait_Analysis’, sheet 6) were used to draw fruiting body, slug and spore dimensions at the correct relative sizes onto a schematic of the deep topology of the Dictyostelid phylogeny. Only stalks are presented at one-third of their length, relative to diameter. For all categorical characters that showed a well-defined character history, character states at major nodes were retrieved from the electronic supplementary material, figure S6 and plotted as cartoons onto the phylogeny.