Fig. 4.
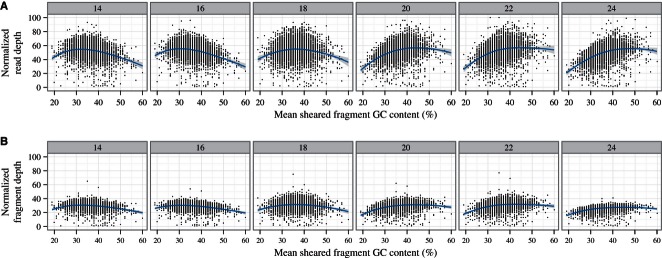
Read depths are influenced by GC content of sheared fragments and number of PCR cycles. PCR cycles vary from 14 (left) to 24 (right) in six separate C. elegans samples (Replicate 2 samples used, see Table S1, Supporting Information). Caenorhabditis elegans RAD loci with restriction fragment lengths over 10 kb are shown, to minimize restriction fragment length bias. (A) Normalized read depth for mean sheared fragment GC content is shown with Loess curve. (B) Removing PCR duplicates (collapsing multiple copies of unique read pairs) approximates number of sheared fragments and partially corrects PCR bias.
