Abstract
In invertebrate-parasite systems, the likelihood of infection following parasite exposure is often dependent on the specific combination of host and parasite genotypes (termed genetic specificity). Genetic specificity can maintain diversity in host and parasite populations and is a major component of the Red Queen hypothesis. However, invertebrate immune systems are thought to only distinguish between broad classes of parasite. Using a natural host-parasite system with a well-established pattern of genetic specificity, the crustacean Daphnia magna and its bacterial parasite Pasteuria ramosa, we found that only hosts from susceptible host-parasite genetic combinations mounted a cellular response following exposure to the parasite. These data are compatible with the hypothesis that genetic specificity is attributable to barrier defenses at the site of infection (the gut), and that the systemic immune response is general, reporting the number of parasite spores entering the hemocoel. Further supporting this, we found that larger cellular responses occurred at higher initial parasite doses. By studying the natural infection route, where parasites must pass barrier defenses before interacting with systemic immune responses, these data shed light on which components of invertebrate defense underlie genetic specificity.
Keywords: Daphnia magna, genetic specificity, invertebrate immunity, Pasteuria ramosa
INTRODUCTION
For invertebrate hosts, the probability of becoming infected following exposure to a parasite is often dependent on the specific pairing of host and parasite genotypes. This is genetic specificity, and it manifests statistically as a host genotype-by-parasite genotype interaction (Carius et al. 2001; Schmid-Hempel and Reber 2004; Wilfert and Schmid-Hempel 2008). Genetic specificity is a mechanism for the maintenance of host and parasite diversity in natural populations because it can lead to negative frequency-dependent selection (Hamilton 1980; Byers 2005). However, invertebrate immune systems are thought to vary in response only to very broad classes of parasites and pathogens, e.g. between Gram positive and Gram negative bacteria (Lemaitre et al. 1997; Ferrandon et al. 2003). If invertebrate immune systems cannot distinguish between individual parasite strains, how can genetic specificity for infection outcome be so widespread?
Many studies of invertebrate defense involved the injection of pathogen mimics directly into the hemocoel. While this has shed a bright light on general immune responses, much can also be learned by studying a natural infection process where living parasites are tested against all host defences, including barriers and systemic responses (Little et al. 2005; Parker et al. 2011). For example, Duneau et al. (2011) studied the natural infection process that occurs when the specialist, trophically transmitted bacteria Pasteuria ramosa invades its host, the crustacean Daphnia magna. They provided evidence that infection success was determined at the gut wall, prior to contact with the systemic immune system. Specifically, they showed that the pathogen could attach to the oesophagus of a susceptible host genotype, but not to the esophagus of a resistant genotype (Duneau et al. 2011).
The Daphnia magna-Pasteuria ramosa system shows some the very strongest patterns of genetic specificity yet observed (Carius et al. 2001; Luijckx et al. 2011), but much remains to be determined about the mechanisms. Here, we sought to gain further insight into specificity in this system by testing whether the immune response played the role of parasite-killer in resistant genotypes, influenced variation in virulence among susceptible genotypes, or whether the immune response only sometimes plays a role, say, in certain host genotypes, against certain parasite genotypes, or at certain doses.
We exposed multiple genotypes of Daphnia magna to a fixed dose of multiple genotypes of Pasteuria ramosa. In a second experiment, we exposed host genotypes to a varying dose of a single P. ramosa genotype. Following parasite exposure, we documented infection status (infected or not), measures of host and parasite fitness and the magnitude of the induced host cellular response (a measure of host systemic immune activity). Thus, we were able to link a measure of host immune function with infection outcome (and thus host fitness potential), when the parasite was allowed to infect via the natural route.
MATERIAL AND METHODS
Study organisms
Daphnia magna is a small freshwater crustacean. It is cyclically parthenogenetic, and usually reproduces asexually, but has sex under stressful conditions (Kleiven et al. 1992). By keeping Daphnia in good conditions, it is therefore possible to maintain independent clonal lineages. Pasteuria ramosa is an obligate bacterial endoparasite of Daphnia, and its transmission spores are ingested when the host is filter feeding. Once inside the gut, these spores infect and undergo development in the host’s hemolymph, ultimately causing host sterilization (Ebert et al. 1996). The next generation of parasite transmission spores are released from host cadavers (Ebert et al. 1996).
Hemocytes are known to be an important immune defense in many invertebrates (Ataev and Coustau 1999; Elrod-Erickson et al. 2000; Kraaijeveld et al. 2001; Canesi et al. 2002; Cotter et al. 2004). Previous work has documented two types of hemocyte circulating in the Daphnia hemolymph (named granulocytes and plasmatocytes) (Metchnikoff 1884; Auld et al. 2010), and that the number of circulating plasmatocytes increases substantially 4-6 hours after exposure P. ramosa in some host genotypes, but not others (Auld et al. 2010). In the present study, we record the number of circulating plasmatocytes in control and P. ramosa-exposed hosts in order to test for an induced cellular response.
We used eight Daphnia genotypes (=clones), named GG4, GG16, GG17, GG18, GG20, GG22, GG23 and GG26 and five P. ramosa isolates, named Sp1, Sp8, Sp13, Sp17, Sp23. All hosts and parasites originated from a pond in Gaarzerfeld, Germany and were collected in 1997 (Carius et al. 2001). Hosts GG4 and parasites Sp1 and Sp8 were isolated in 1997. These hosts have since been kept in a state of clonal reproduction, and the parasite spores were frozen at −20°C. The remaining host genotypes were hatched from resting eggs (ephippia) in 2009 (Auld et al. 2010), and parasite isolates Sp17 and Sp23 were obtained by exposing GG17 and GG23 to the original sediment collected from Gaarzerfeld. A single infected GG17 and GG23 were then randomly chosen and each were homogenised in 5 ml of ddH2O to make spore suspensions. These were propagated using Daphnia of the same original genotype.
Experimental setup
Both experiments followed similar protocols. Eight-to-twelve replicates of each host genotype were kept for three generations to minimise variation in maternal effects. A replicate consisted of three jars, each containing five Daphnia and 200 ml of artificial Daphnia medium (Kluttgen et al. 1994). Daphnia were fed 1 ABS of chemostat-grown Chlorella vulgaris algal cells per Daphnia per day, (ABS refers to optical absorbance of 650 nm white light by the C. vulgaris culture), and all jars were incubated at 20°C on a 12:12 hour light/dark cycle. Medium was refreshed three times per week, or after the Daphnia had a clutch of offspring. Offspring from each maternal replicate were divided between parasite treatments for each of the two experiments. Ultimately, there were five Daphnia per replicate jar. These Daphnia were kept in the same conditions as maternal replicates until three of the five Daphnia in each jar had deposited eggs in their brood pouches; when this happened they were ready for exposure to their experimental treatments.
For the specificity experiment, host genotypes GG4, GG16, GG18, GG23 and GG26 were exposed to six parasite treatments: 5 × 105 spores (in 100 μl) of either Sp1, Sp8, Sp13, Sp17 and Sp23, or to a no-parasite control (consisting of 100 μl of homogenised healthy Daphnia). There were 10 replicates of each host-parasite combination. The Daphnia from each replicate were placed in a well of a 24-well plate (Costar, Corning Inc., NY) with 1 ml of artificial medium and 100 μl of their designated parasite treatment (i.e. one of parasite isolates, or the no-parasite control treatment). After five hours of exposure to the parasite, Daphnia were washed in artificial medium and four of the Daphnia from each replicate were dried on a paper towel, placed on a Petri dish and their hearts were pierced with a 25 gauge needle (BD Microlance, Drogheda, Ireland). 1.0 μl of hemolymph was pipetted from each and mixed with 4 μl of anticoagulant buffer (98 mM NaOH, 186 mM NaCl, 17 mM EDTA and 41 mM citric acid, pH adjusted to 4.5) (Lavine et al. 2005), giving a total of 8 μl of hemolymph solution. 2 μl of the resulting solution was placed in a fertility counting chamber [0.001 mm2×0.100 mm (depth)] (Hawksley, Lancing, Sussex, UK), and the density of circulating plasmatocytes (henceforth hemocytes) per Daphnia was determined.
The fifth Daphnia from each replicate was placed in a jar with 60 ml of artificial medium and maintained under the same conditions as the maternal replicates. All Daphnia were checked daily for reproduction and mortality. If they had a clutch, the age of reproduction (in days) and the number of offspring was recorded. Offspring and dead hosts were removed from the jars and dead hosts were checked for evidence of P. ramosa infection by pressing them under a cover slip and examining them under a transmission microscope. The experiment was terminated on day 40, when all surviving hosts were frozen at −20°C. Counts of P. ramosa transmission spores were made from each host surviving until the end of the experiment by homogenizing individual Daphnia in 100 μl of ddH2O. Two independent counts of spores were made from the resulting suspension in a Neubauer (Improved) counting chamber (0.0025 mm2 × 0.1 mm depth).
For the dose experiment, host genotypes GG4, GG16, GG17, GG18, GG20, GG22 were exposed to 5 doses of parasite strain Sp1: 10, 102, 103, 104, 106 spores in 100 μl, or to a no parasite control (again, 100 μl of homogenised healthy Daphnia). There were 8-12 replicates per host-dose combination. Four of the Daphnia per replicate jar were used to determine hemocyte densities and the fifth Daphnia was used to collect host fecundity, host mortality and transmission spore count data following protocols described earlier.
Analysis
Analyses were similar for both experiments, and were performed using R (R 2005). The proportion of infected hosts was analysed by fitting a GLM with a quasibinomial error structure (to control for overdispersion) to data from parasite-exposed treatments, and the number of circulating hemocytes was analysed by fitting an ANOVA to all data. For the specificity data, host genotype, parasite genotype and their interaction were included as explanatory variables. For the dose data, the explanatory variables were host genotype, Log10[dose] and their interaction. For both data sets, host reproduction was analysed by testing for a difference between offspring counts from infected and healthy individuals using a Welch’s two-sample t-test.
As expected from this study population, the specificity experiment found a significant host genotype-by-parasite genotype interaction for infection success, and three categories were clearly evident: infective host-parasite combinations, non-infective host-parasite combinations and controls. We tested for differences in hemocyte densities between these categories using a one-way ANOVA. Inspection of the hemocyte count data from the dose experiment suggested a non-linear relationship between hemocyte density and initial parasite dose, so we analysed hemocyte density using a general linear model with host genotype, Log10[dose], (Log10[dose])2 and all two-way interactions as explanatory variables.
The number of parasite transmission spores was analysed by fitting a type III ANOVA to spore counts from infected hosts only. For specificity data, host reproduction, hemocyte density, host genotype and parasite genotype fitted as explanatory variables. Mortality during the experiment meant that only a fraction of infected hosts survived until the end of the experiment (day 40), and thus spore counts were made from 24 infected hosts. In the dose experiment, only host genotype GG4 suffered infections at all parasite doses, so analysis was restricted to this genotype; Log10[hemocyte density], Log10[dose], (Log10[dose])2 and all two-way interactions were fitted as explanatory variables.
RESULTS
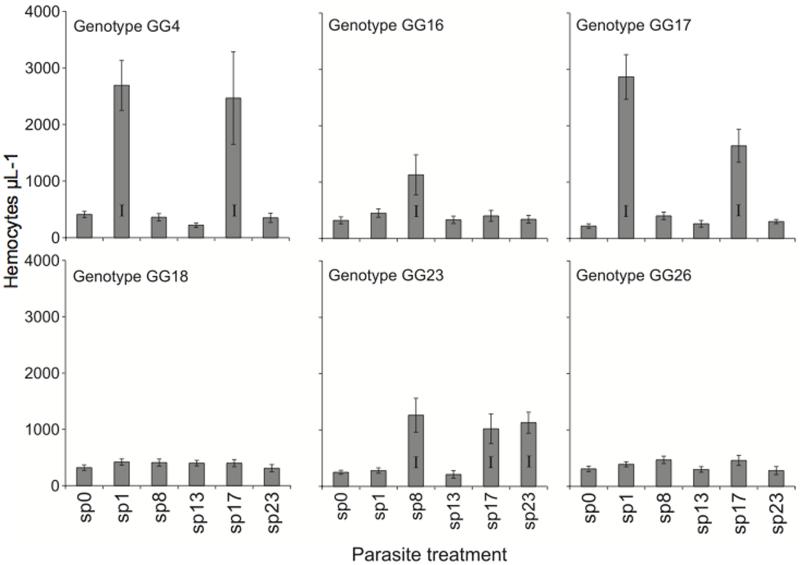
For the genetic specificity experiment, the proportion of hosts becoming infected ranged from 0 to 70%, but whether infection occurred at all was strongly dependent on the host-parasite genotypic combination (Fig. 1, Table 1), confirming earlier work (Carius et al. 2001; Luijckx et al. 2011). The fitness impacts of infection were substantial: infected hosts had 30.6 ±1.4 offspring, whereas healthy hosts had 83.9 ± 1.1 (t71 = 29.27, p < 0.0001). These infection outcomes were mirrored by hemocyte increases. In particular, parasite exposure led to elevated hemocyte counts in susceptible host-parasite combinations, but in combinations that never resulted in infections, exposure to parasites did not change hemocyte densities compared to unexposed controls (Fig. 1, Table 1).
Figure 1.
Hemocyte counts (± 1 s.e.) and infection success for multiple host genotype-parasite strain combinations. Combinations with successful infections are denoted with an I.
Table 1.
Summary of analyses of the proportion of infected hosts following parasite exposure, number of circulating hemocytes five hours after parasite exposure and number of parasite transmission spores at the end of the experiments.
| Proportion of infected hosts |
Number of hemocytes |
Number of parasite spores |
|
|---|---|---|---|
| Genetic specificity experiment |
|||
| Host genotype | F5,299 = 26.17 *** | F5,278 = 20.78 *** | F314 = 11.84 ** |
| Parasite genotype | F4,299 = 14.78 *** | F4,278 = 31.92 *** | F4,14 = 10.79 ** |
| Host × parasite | F20,299 = 4.60 *** | F20,278 = 8.67 *** | - |
| Host fecundity | - | - | F1.14 = 0.63 |
| Host hemocytes | - | - | F1,14 = 2.83 |
| Dose experiment | |||
| Host genotype | F5,333 = 34.64 *** | F5,380 = 0.46 | - |
| Log10[dose] | F1,333 = 32.24 *** | F1,380 = 74.90 *** | F1,26 = 4.34 * |
| (Log10[dose])2 | - | F1333 = 40.95 *** | - |
| Host × log10[dose] | F5333 = 1.86 | F5,380 = 14.12 *** | - |
| Host × (log10[dose])2 | - | F5,380 = 8.61 *** | - |
| Log10[hemocytes] | - | - | F1,26 = 0.66 |
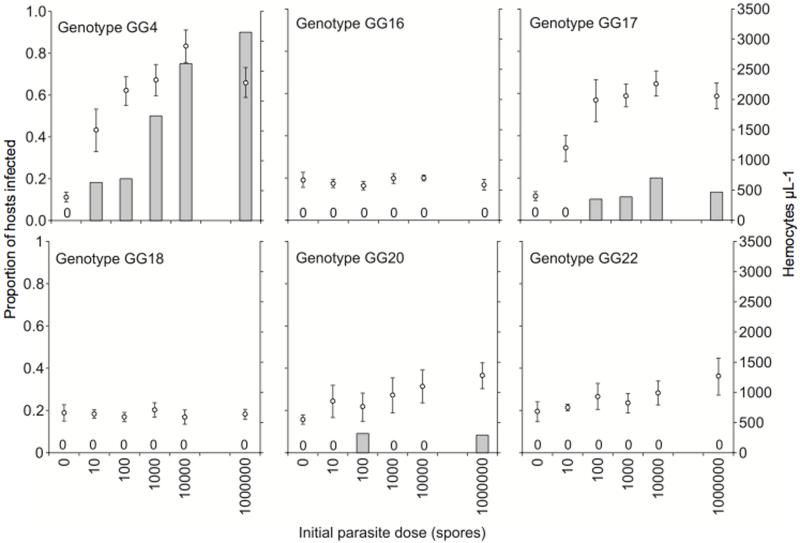
For the dose experiment, the proportion of hosts that became infected ranged from 0 to 90%, and once again, infected hosts had fewer offspring (34.4 ± 1.9) than their healthy counterparts (77.6 ± 1.1: t63 = 19.70, p < 0.0001). Increasing the initial dose of parasites in a susceptible combination served to increase both infection success and the magnitude of cellular response (Fig. 2, Table 1). The number of hemocytes levelled off at high parasite doses in the two most susceptible host genotypes (GG4 and GG17).
Figure 2.
Proportion of infected hosts and number of circulating hemocytes (±1 s.e.) over six parasite doses. Cellular responses were only documented in host genotypes that went on to suffer infections. Treatments that did not result in infections are denoted with a 0.
DISCUSSION
In this study, we sought to link the induced host cellular response (a systemic immune response) with infection success in a host-parasite system that exhibits strong genetic specificity for infection. Our data, combined with those of Duneau et al (2011) suggest the presence and magnitude of induced cellular response appears to report the number of parasite spores passing from the gut to the hemocoel: the presence of an induced cellular response perfectly predicted which host-parasite genotype combinations were susceptible (Fig. 1) and the magnitude of the cellular response increased with initial parasite dose, directly mirroring the likelihood of infection (Fig. 2). Our findings therefore indicate that the mechanisms that determine host susceptibility under genetic specificity operate before the parasite can elicit a substantial host cellular response. This may involve factors expressed at the gut epithelium, either proteins constitutively expressed by host and parasite that allow the parasites to gain entry, or a localised (and presumably rapid) epithelial immune response.
Other studies add mechanistic support to the hypothesis that infection success is determined before the parasite reaches the host’s hemocoel: Duneau et al. (2011) observed attachment of P. ramosa spores on the esophagus of a susceptible D. magna genotype, but a distinct lack of attachment in a resistant genotype. A related parasite, P. penetrans, infects its nematode hosts by attaching to the heparin-binding domain of the host cuticle (Sayre and Starr 1985; Mohan et al. 2001; Schmidt et al. 2008). To persist in the host population, such receptors would have evolved for another important purpose, perhaps for the recognition of nutrients, symbionts or commensal bacteria, and certain parasite strains may have taken advantage of these receptors by expressing chemical features that imitate food or commensal bacteria - ‘wolves in sheep’s clothing’. The ability of a host to recognize and eliminate a parasite may depend on a lock-and-key mechanism where each receptor must effectively bind to a molecular feature on the parasite. Thus, host-parasite coevolution may occur through shifting allele frequencies in host receptors and matching parasite molecular features, with little involvement of systemic host immune responses.
While the barrier hypothesis has merit, we cannot exclude the possibility that genetic specificity stems from other immune responses or constitutively expressed components of the immune system. Although there are substantially more hemocytes in susceptible host-parasite genotypic combinations, (Fig 1) those hemocytes may, for example, be less phagocytically active or fail to release immune cytotoxins (Nappi and Ottaviani 2000). However, recent studies have examined the expression of candidate immune genes in D. magna in unexposed and in P. ramosa-exposed hosts, 0h 1h, 2h, 4h, 8h, 12h and 24h after treatment exposure (Labbé and Little 2009; Labbé et al. 2009; Decaestecker et al. 2011). These studies found no major parasite-induced changes in gene expression for alpha-2-macroglobulin, propenoloxidase, and two nitric oxide synthase genes, and these genes are therefore unlikely to form part of the Daphnia defense against P. ramosa. To more fully exclude the 4 possibility that rapid systemic host immune responses prevent successful P. ramosa infections, or that P. ramosa switches off host immune responses, it would be helpful to compare the transcriptomes of parasite-exposed and resistant, parasite-exposed and healthy and unexposed D. magna.
On the whole, early assertions, based on the study of systemic immunological mechanisms, that the invertebrate immune response is rather general and possesses the capacity to distinguish only broad classes of pathogens (say, fungal versus bacterial) are not disproved in this instance – although there is often considerable host genetic variation for the presence and magnitude of such responses (Lazzaro et al. 2004; Auld et al. 2010) that can also depend on interactions with the environment (e.g. Seppälä and Jokela 2010). Revealing the mechanisms of genetic specificity clearly requires consideration that infection is a process that includes a series of steps (Auld et al. 2010; Duneau et al. 2011), including that pathogens must overcome constitutive barriers, epithelial responses (Boulanger et al. 2004; Bosch et al. 2009; Riddell et al. 2009), and systemic responses (Haine et al. 2008). Much work on invertebrate immunity has injected live pathogens, dead pathogens or pathogen mimics directly into the hemocoel of hosts, thus limiting the opportunity to study important mechanisms at other steps of the infection process. Consideration of the complete infection route thus reveals what is potentially a broad distinction between vertebrate and invertebrate defense systems, in particular that in vertebrate defense, barriers are general and specificity originates with the developing systemic immune response, while for invertebrates, specificity lies with barrier mechanisms.
Such a difference between vertebrate and invertebrate anti-parasite defenses may stem from their different body plans. Invertebrates have an open circulatory system, so an infecting parasite has access to many different body tissues once inside the host’s hemocoel, but the closed nature of the vertebrate circulatory system makes it easier for the host to isolate and destroy an infecting parasite before it reaches other tissues. Invertebrates may therefore have been under stronger selection to keep parasites out in the first place, and thus mount their most potent defenses at the barrier.
Genetic specificity attracts considerable research effort because it has profound implications for host-parasite coevolution (Lambrechts 2010). In particular, as each parasite strain can select against only a subset of hosts and vice versa, this fosters negative frequency-dependent selection (Jaenike 1978; Hamilton 1980; Byers 2005), which is a mechanism for the maintenance of polymorphism, and a key component of the Red Queen hypothesis (Hamilton 1980). The occurrence of genetic specificity is therefore linked to one of the most fundamental and challenging questions in evolutionary biology: what is the adaptive significance of sexual reproduction? Genetic specificity is possibly the norm in invertebrate host-parasite interactions (Wilfert and Schmid-Hempel 2008), and yet the mechanisms underlying genetic specificity are not adequately understood (Lambrechts 2010). There is considerable knowledge from, for example, studies of transcriptional regulation or gene knockdown experiments about genes that play a role in infection (Vallet-Gely et al. 2008; Irazoqui et al. 2010), but we remain in the dark about the genes that underlie the genetic polymorphisms of defense. By highlighting the likely physical location of genetic specificity in the Daphnia-Pasteuria system, the present study and others (Riddell et al. 2009; Duneau et al. 2011) can hopefully advance the search for these mechanisms and isolate the arena in which host-parasite coevolution occurs
ACKNOWLEDGEMENTS
SKJRA was supported by a NERC Ph.D. studentship. TJL was supported by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Sciences. KHE was supported by the Erasmus Foundation. Comments from N. Gerardo, S. Nuismer and an anonymous reviewer improved this manuscript.
REFERENCES
- Ataev GL, Coustau C. Cellular response to Echinostoma caproni infection in Biomphalaria glabrata strains selected for susceptibility/resistance. Developmental and Comparative Immunology. 1999;23:187–198. doi: 10.1016/s0145-305x(99)00023-3. [DOI] [PubMed] [Google Scholar]
- Auld SKJR, Scholefield JA, Little TJ. Genetic variation in the cellular response of Daphnia magna (Crustacea: Cladocera) to its bacterial parasite. Proceedings of the Royal Society of London Series-B Biological Sciences. 2010;277:3291–3297. doi: 10.1098/rspb.2010.0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, Stanisak M, Groetzinger J, Jung S, Podschun R, Bartels J, Harder J, Schroeder J-M. Uncovering the evolutionary history of innate immunity: The simplest metazoan Hydra uses epithelial cells for host defence. Developmental and Comparative Immunology. 2009;33 doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Boulanger N, Lowenberger C, Volf P, Ursic R, Sigutova L, Sabatier L, Svobodova M, Beverley SM, Spaeth G, Brun R, Pesson B, Bulet P. Characterization of a Defensin from the Sand Fly Phlebotomus duboscqi Induced by Challenge with Bacteria or the Protozoan Parasite Leishmania major. Infection and Immunity. 2004;72:7140–7146. doi: 10.1128/IAI.72.12.7140-7146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers DL. Evolution in heterogeneous environments and the potential of maintenance of genetic variation in traits of adaptive significance. Genetica. 2005;123:107–124. doi: 10.1007/s10709-003-2721-5. [DOI] [PubMed] [Google Scholar]
- Canesi L, Gallo G, Gavioli M, Pruzzo C. Bacteria-hemocyte interactions and phagocytosis in marine bivalves. Microscopy Research and Technique. 2002;57:469–476. doi: 10.1002/jemt.10100. [DOI] [PubMed] [Google Scholar]
- Carius HJ, Little TJ, Ebert D. Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution. 2001;55:1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. [DOI] [PubMed] [Google Scholar]
- Cotter SC, Kruuk LEB, Wilson K. Costs of resistance: genetic correlations and potential trade-offs in an insect immune system. Journal of Evolutionary Biology. 2004;17:421–429. doi: 10.1046/j.1420-9101.2003.00655.x. [DOI] [PubMed] [Google Scholar]
- Decaestecker E, Labbé P, Ellegaard K, Allen JE, Little TJ. Candidate innate immune system gene expression in the ecological model Daphnia. Developmental and Comparative Immunology. 2011;35:1066–1075. doi: 10.1016/j.dci.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duneau D, Luijckx P, Ben-Ami F, Laforsch C, Ebert D. Resolving the infection process reveals striking differences in the contribution of environment, genetics and phylogeny to host-parasite interactions. BMC Biology. 2011;9:11. doi: 10.1186/1741-7007-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Rainey P, Embley TM, Scholz D. Development, life cycle, ultrastructure and phylogenetic position of Pasteuria ramosa Metchnikoff 1888: Rediscovery of an obligate endoparasite of Daphnia magna Straus. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 1996;351:1689–1701. [Google Scholar]
- Elrod-Erickson M, Mishra M, Schneider D. Interactions between the cellular and humoral immune responses in Drosophila. Current Biology. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hoffmann JA. Sensing infection in Drosophila: Toll and beyond. Seminars in Immunology. 2003;16:43–53. doi: 10.1016/j.smim.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Haine ER, Y. M, Siva-Jothy MT, Rolff J. Antimicrobial defense and persistent infection in insects. Science. 2008;21:1257–1259. doi: 10.1126/science.1165265. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. [Google Scholar]
- Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from C. elegans and primitive invertebrates. Nature Reviews Immunology. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J. A hypothesis to account for the maintenance of sex within populations. Evolutionary Theory. 1978;3:191–194. [Google Scholar]
- Kleiven OT, Larsson P, Hobaek A. Sexual reproduction in Daphnia magna requires three stimuli. Oikos. 1992;65:197–206. [Google Scholar]
- Kluttgen BU, Dulmer U, Engels M, Ratte HT. ADaM, an artificial freshwater for the culture of zooplankton. Water Research. 1994;28:743–746. [Google Scholar]
- Kraaijeveld AR, Limentani EC, Godfray HCJ. Basis of the trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Proceedings of the Royal Society of London Series B Biological Sciences. 2001;268:259–261. doi: 10.1098/rspb.2000.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé P, Little TJ. ProPhenolOxidase in Daphnia magna: cDNA sequencing and expression in relation to resistance to pathogens. Developmental and Comparative Immunology. 2009;33:674–680. doi: 10.1016/j.dci.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Labbé P, McTaggart SJ, Little TJ. An ancient immunity gene duplication in Daphnia magna: RNA expression and sequence analysis of two nitric oxide synthase genes. Developmental and Comparative Immunology. 2009;33:1000–1010. doi: 10.1016/j.dci.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L. Dissecting the genetic architecture of host-pathogen specificity. PLoS Pathogens. 2010;6:e1001019. doi: 10.1371/journal.ppat.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine MD, Chen G, Strand MR. Immune challenge differentially affects transcript abundance of three antimicrobial peptides in hemocytes from the moth Pseudoplusia includens. Insect Biochemistry and Molecular Biology. 2005;35:1335–1346. doi: 10.1016/j.ibmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proceedings of the National Academy of Sciences of the USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TJ, Hultmark D, Read AF. Invertebrate immunity and the limits of mechanistic immunology. Nature Immunology. 2005;6:651–654. doi: 10.1038/ni1219. [DOI] [PubMed] [Google Scholar]
- Luijckx P, Ben-Ami F, Mouton L, Du Pasquier L, Ebert D. Cloning of the unculturable parasite Pasteuria ramosa and its Daphnia host reveals extreme genotype-genotype interactions. Ecology Letters. 2011;14:125–131. doi: 10.1111/j.1461-0248.2010.01561.x. [DOI] [PubMed] [Google Scholar]
- Metchnikoff I. A disease of Daphnia caused by a yeast. In: Brock T, editor. A contribution to the theory of phagocytes as agents for attack on disease-causing organisms. Milestones in Microbiology. American Society for Microbiology; Washington D.C: 1884. pp. 132–138. [Google Scholar]
- Mohan S, Fould S, Davies KG. The interaction between the gelatin-binding domain of fibronectin and the attachment of Pasteuria penetrans endospores to nematode cuticle. Parasitology. 2001;123:271–276. doi: 10.1017/s0031182001008411. [DOI] [PubMed] [Google Scholar]
- Nappi AJ, Ottaviani E. Cytotoxicity and cytotoxic molecules in invertebrates. BioEssays. 2000;22:469–480. doi: 10.1002/(SICI)1521-1878(200005)22:5<469::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Parker BJ, Barribeau SM, Laughton AL, de Roode JC, Gerardo NM. Non-immunological defense in an evolutionary framework. Trends in Ecology and Evolution. 2011;26:242–248. doi: 10.1016/j.tree.2011.02.005. [DOI] [PubMed] [Google Scholar]
- R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2005. [Google Scholar]
- Riddell C, Adams S, Schmid-Hempel P, Mallon EB. Differential expression of immune defences is associated with specific host-parasite interactions in insects. PLoS One. 2009;4:e7621. doi: 10.1371/journal.pone.0007621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre RM, Starr MP. Pasteuria penetrans (ex Thorne 1940) nom. rev., comb. n., sp. n., a mycelial endospore-forming bacterium parasitic in plant-parasitic nematodes. Proceedings of the Helminthological Society of Washington. 1985;52:149–165. [Google Scholar]
- Schmid-Hempel P, Reber C. The distribution of genotypes of the trypanosome parasite Crithidia bombi in populations of its host, Bombus terrestris. Parasitology. 2004;129:147–158. doi: 10.1017/s0031182004005542. [DOI] [PubMed] [Google Scholar]
- Schmidt LM, Mouton L, Nong G, Ebert D, Preston J. Genetic and immunological comparison of the cladoceran parasite Pasteuria ramosa with the nematode parasite Pasteuria penetrans. Applied and Environmental Microbiology. 2008;74:259–264. doi: 10.1128/AEM.01778-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä O, Jokela J. Maintenance of genetic variation in immune defence of a freshwater snail: role of environmental heterogeneity. Evolution. 2010;64:2397–2407. doi: 10.1111/j.1558-5646.2010.00995.x. [DOI] [PubMed] [Google Scholar]
- Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nature Reviews Microbiology. 2008;6:302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Schmid-Hempel P. The genetic architecture of susceptibility to parasites. BMC Evolutionary Biology. 2008;8:187–195. doi: 10.1186/1471-2148-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]