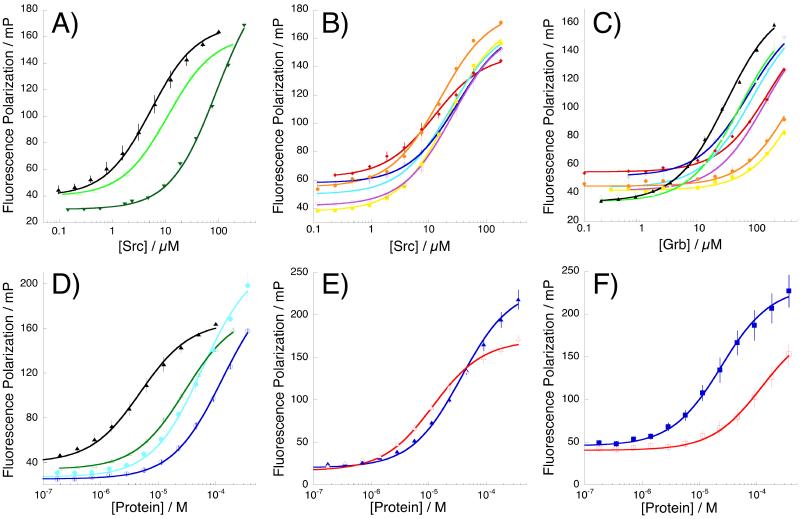
Figure 5.
Representative binding isotherms, showing data and comparisons of key arginine-mimetic peptides. mP = fluorescence polarization in millipolarization units (= polarization/1000). Error bars indicate standard error. (A) hAMI(Arg) (black triangles), hAMI(Dap-Ac) (dark green inverted triangles), and hAMI(gGly) (light green open triangles) binding to Src; (B) α-substituted hAMI arginine mimetic peptides binding to Src: hAMI(gl-Val) (magenta open squares), hAMI(gd-Val) (yellow closed squares), hAMI(gl-Phe) (cyan open circles), hAMI(gd-Phe) (orange closed circles), hAMI(gl-Trp) (blue open diamonds), hAMI(gd-Trp) (red closed diamonds); (C) α-substituted hAMI arginine mimetic peptides binding to Grb: hAMI(Arg) (black closed triangles); hAMI(gGly) (light green open triangles); hAMI(gl-Val) (magenta open squares), hAMI(gd-Val) (yellow closed squares), hAMI(gl-Phe) (cyan open circles), hAMI(gd-Phe) (orange closed circles), hAMI(gl-Trp) (blue open diamonds), hAMI(gd-Trp) (red closed diamonds); (D) hAMI(Arg) binding to Src (closed black triangles) and Grb (open green triangles); AMI(gl-Phe) binding to Src (closed cyan circles) and Grb (open blue circles); (E) hAMII(Arg) binding to Src (closed blue triangles) and Grb (open red triangles); (F) hAMII(gl-Trp) binding to Src (closed blue squares) and Grb (open red squares).