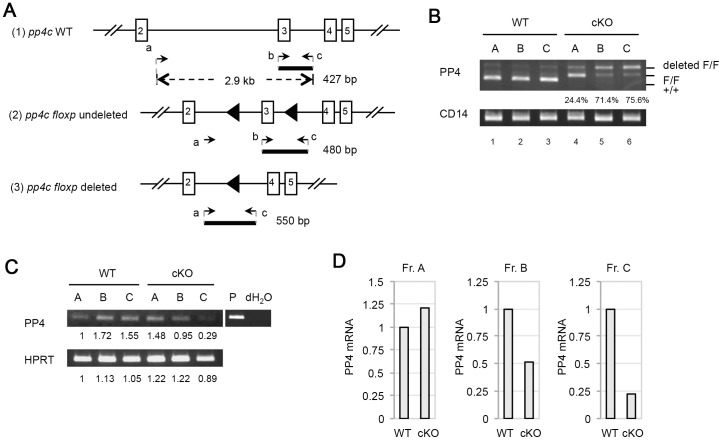
Figure 2. Efficiency of pp4c deletion in B cells.
(A) Schematic illustration of the targeting strategy used to delete pp4c and the primers designed to detect deletion efficiency. For genomic PCR, forward primers “a” (5′-ACGTGATTTGCGAAAGCCTCTCA-3′) and “b” (5′-CTTGGTAGAAGAGAGCAACGTGCAG-3′), and reverse primer “c” (5′-TGCTCTGGTGGCAGGAGATGTGTG-3′), were used as indicated. The PCR products of the WT pp4c allele (427 bp; 1); the pp4c floxp allele before cre-mediated deletion (480 bp; 2); and the pp4c floxp allele after cre-mediated deletion (550 bp; 3) are shown. (B) PCR analysis of genomic DNA from Frs. A, B and C cells from WT and cKO mice (n = 1/group) showing products representing the WT pp4c allele (+/+), the pp4c floxp allele (F/F), and the deleted pp4c allele (deleted F/F). Percentages shown are deletion efficiencies. CD14, loading control. Results are representative of three independent experiments. (C) RT-PCR analysis of PP4 and HPRT mRNAs in Frs. A, B and C cells from WT and cKO mice (n = 2/group). Numbers are the relative mRNA levels quantified by Image J and normalized to the WT Fr. A value (set to 1). P, positive control for PP4. HPRT, loading control. Results are representative of three independent experiments. (D) Quantitation of the mRNA levels in the cells in (C) after normalization to HPRT values.