Abstract
The hickory genus (Carya) contains ca. 17 species distributed in subtropical and tropical regions of eastern Asia and subtropical to temperate regions of eastern North America. Previously, the phylogenetic relationships between eastern Asian and eastern North American species of Carya were not fully confirmed even with an extensive sampling, biogeographic and diversification patterns had thus never been investigated in a phylogenetic context. We sampled 17 species of Carya and 15 species representing all other genera of the Juglandaceae as outgroups, with eight nuclear and plastid loci to reconstruct the phylogeny of Carya. The phylogenetic positions of seven extinct genera of the Juglandaceae were inferred using morphological characters and the molecular phylogeny as a backbone constraint. Divergence times within Carya were estimated with relaxed Bayesian dating. Biogeographic analyses were performed in DIVA and LAGRANGE. Diversification rates were inferred by LASER and APE packages. Our results support two major clades within Carya, corresponding to the lineages of eastern Asia and eastern North America. The split between the two disjunct clades is estimated to be 21.58 (95% HPD 11.07-35.51) Ma. Genus-level DIVA and LAGRANGE analyses incorporating both extant and extinct genera of the Juglandaceae suggested that Carya originated in North America, and migrated to Eurasia during the early Tertiary via the North Atlantic land bridge. Fragmentation of the distribution caused by global cooling in the late Tertiary resulted in the current disjunction. The diversification rate of hickories in eastern North America appeared to be higher than that in eastern Asia, which is ascribed to greater ecological opportunities, key morphological innovations, and polyploidy.
Introduction
As one of the typical phytogeographic disjunctions in the Northern Hemisphere, the eastern Asian (EA)-eastern North American (ENA) floristic disjunction pattern has received considerable attention (see 1-4 and references therein). This pattern is often explained by the boreotropical flora hypothesis [5]. A relatively continuous, homogenous mesophytic forest that spanned the Northern Hemisphere during the climatically warm mid-Tertiary became fragmented as global temperature cooled down in the late Tertiary and Quaternary [1,3,5-9]. Both the Bering land bridge (BLB) [10,11] and the North Atlantic land bridge (NALB) [12,13] probably contributed to the floristic intercontinental exchanges to form the boreotropical flora. Paleontological and molecular data suggest that BLB was used mostly by temperate taxa during the late Miocene and Pliocene (<10 Ma [4,8,14]). NALB has been viewed as a crucial route for the spread of subtropical and tropical taxa in the early Tertiary [5,6,13,15]. So far, most studies have focused on the temperate taxa in EA and ENA (see 1,3,14,16 and references therein); our understanding of the biogeographic relationships of the groups involving subtropical and tropical elements is still preliminary [3]. It is also noteworthy that species diversities within the two regions differ markedly. For taxa that distinctly distributed in EA and ENA, the former region usually has higher species richness than the latter [1,17,18]; this has been attributed to greater net speciation rate and accelerated molecular evolution in EA species [19]. However, several groups present a reverse biodiversity pattern, such as Catalpa (Bignoniaceae), Lyonia (Ericaceae), and Carya (Juglandaceae), in which ENA harbors more species [1]. Thus, to better understand the evolution of biodiversity in the two regions, an effort should be made to broaden the phylogenetic and biogeographic investigations on various disjunct taxa.
The hickory genus Carya, one of eight genera of the Juglandaceae, exhibits an intercontinentally disjunct distribution between EA and ENA (Figure 1). According to the records in Flora of North America [20] and Flora of China [21], Carya contains eleven species in ENA and four in EA. The ENA Carya ovalis (Wangenh.) Sarg. was treated as a synonym of C . glabra by Flora of North America [20], but this treatment is still controversial as both taxa quite distinct morphologically [22]. Carya sinensis Dode, endemic to southwestern China, was formerly treated as the monotypic genus Annamocarya, but it belongs to Carya according to molecular phylogenetic analyses [23,24]. Carya poilanei (Chev.) Leroy is also an accepted name of an EA hickory [25], which only has two specimen collections in Laos, collected in 1932 and 1937 [26]. Using ITS data, Xiang et al. [19] sampled three EA species and six ENA species and found that the EA clade was nested within the ENA grade; thus the two major clades of Carya, corresponding to geographical distribution, were not recovered. Based on morphology (64 characters) in combination with sequence data from three genomic regions (ITS, trnL-F, and atpB-rbcL), sampling the same nine species, two clades corresponding to the distributions of hickories in EA and ENA were recovered [23,24]. Because the importance of increased taxon sampling to enhance phylogenetic accuracy has been supported by several studies (see 27,28 and references therein), it is desired to test the reliability of phylogenetic relationships between EA and ENA species of Carya with an analysis including more species and more molecular loci.
Figure 1. Geographic distribution of both extant and extinct Carya species.
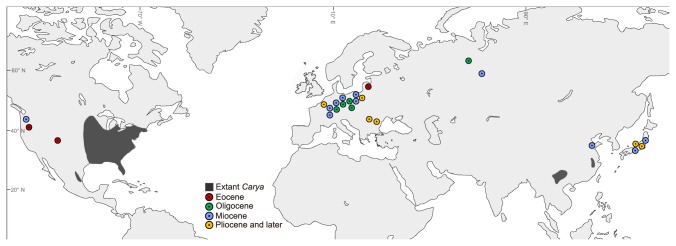
The black areas indicate the geographical distribution of extant Carya showing its disjunction between EA and ENA. Colored circles denote the locations of Carya fruit fossils (detailed information of each fossil in Table S4).
According to the climate classification of Troll and Paffen (see 29), in EA, three hickory species are distributed in subtropical regions ( C . cathayensis , C . hunanensis , and C . kweichowensis ), and three in tropical regions ( C . poilanei , C. sinensis, and C . tonkinensis ). The ENA hickory species are distributed in the subtropical to temperate regions, with several species more prominent in temperate regions, such as C . glabra , C . ovata , and C . illinoinensis . Moreover, Carya also has a rich macrofossil record of fruits and leaves in North America [30,31] and Asia [30], as well as Europe [30,32,33]. Xiang et al. [34,35] have suggested that fossil taxa, especially those from sites outside the extant distribution, should be included in biogeographic analyses. Morphological data can be collected from both extinct and extant taxa, allowing the inclusion of fossils in the analyses and providing a more complete window to the evolutionary history that might not be revealed by molecular data alone [36,37]. The Juglandaceae contains at least eight extinct genera [30,38]. Some studies have attempted to reconstruct the genus-level phylogeny for Juglandaceae including extant and extinct genera [23,24,30]. It is feasible to explore the biogeography of Carya in the phylogenetic framework and geographic range of the whole family. Thus, Carya offers a remarkable opportunity to study biogeography and diversification of an EA-ENA disjunct group distributed across the temperate, subtropical and tropical regions in two continents.
The aims of our study were to (1) investigate phylogenetic relationships between EA and ENA species of Carya using eight plastid (matK, rbcL-atpB, rpoC1, rps16, trnH-psbA, and trnL-F) and nuclear (ITS and phyA) loci and an extensive taxon sampling, (2) reconstruct the historical biogeography of Carya by combining fossil, morphological, and molecular data, and (3) explore the causes of the unusual species richness pattern of EA and ENA Carya.
Materials and Methods
Ethics Statement
No special permits were required for this study because all samples were collected by researchers with introduction letters of IBCAS (Institute of Botany, Chinese Academy of Sciences) in Beijing, and this study did not involve endangered or protected species. Voucher specimens were deposited in the Herbarium, Institute of Botany, Beijing (PE).
Sampling, DNA isolation, and sequencing
We sampled 17 accessions representing five species from EA, including C. sinensis (formerly treated as Annamocarya sinensis ), recorded in Flora of China [21], and eleven species from ENA as recognized in Flora of North America [20] as well as C . ovalis . Only C . poilanei was not sampled because no material was available for no more records have been found since 1937 [26]. Fifteen species representing all other seven genera of Juglandaceae were also sampled. All trees were rooted with Rhoiptelea chiliantha [23,24,39]. Voucher information and GenBank accession numbers are listed in Table S1.
Eight markers, including plastid (matK, rbcL-atpB, rpoC1, rps16, trnH-psbA, and trnL-F) and nuclear (ITS and phyA) loci were used to reconstruct the phylogeny of Carya and its relatives. Total DNA was extracted from silica-gel-dried leaf material using TianGen extraction kits (Beijing, China). The selected DNA regions were amplified with standard polymerase chain reaction (PCR). PCR primers used for amplifying and sequencing the eight plastid and nuclear loci are shown in Table S2.
Molecular phylogenetic analyses
The resulting sequences were initially subjected to a search in BLAST (implemented by the National Center for Biotechnology Information (NCBI) website http://www.ncbi.nlm.nih.gov) against the GenBank nucleotide database to test for contamination and to confirm the targeted markers. All correct sequences were aligned using Clustal X version 2.0 [40], then adjusted manually in BioEdit version 4. 8. 10 [41]. Phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference (BI) methods. ML analyses were done at the Cyperinfrastructure for Phylogenetic Research (CIPRES; www.phylo.org) running RAxML-HPC2 on XSEDE [42,43] with the default settings except that the GTR + Г model was applied. Analyses of each independent data set produced no topological discordance with ML bootstrap proportions > 70%, and the eight data sets were therefore concatenated, yielding a combined molecular data set of 6024 characters (Dataset S1). Substitution model parameters were estimated separately for two partitions as plastid (matK, rbcL-atpB, rpoC1, rps16, trnH-psbA, and trnL-F) and nuclear (ITS and phyA) data partitions. The appropriate substitution models were determined using jModelTest version 1.0 [44]. Both partitions used the GTR + Г model. Bayesian inference was performed using MrBayes version 3.1.2 [45]. The MCMC algorithm was run for 5,000,000 generations with four incrementally heated chains, starting from random trees and sampling one out of every 1000 generations. Trees sampled before stable posterior probability (PP) values had been reached were excluded from consensus as a burn-in phase (initial 20% of the sampled trees), and the remaining trees were used to construct the Bayesian majority-rule consensus tree.
Morphological phylogenetic analyses
A previously published morphological matrix [24] with 64 vegetative, floral, and fruit characters was modified to include more extant and extinct species, and the modified matrix (Table S3) was used to reconstruct a phylogenetic tree including both extant and fossil species of the Juglandaceae. Seven extinct genera have been included, with three fossils, representing extinct genera Hooleya, Palaeocarya, and Paraengelhardia newly added in this study. The coding of their morphological characters of the fossils was done according to the descriptions of Manchester [30]. Combined analysis of fossil and extant taxa with morphological and molecular data was performed using the DNA scaffold method [24] under Maximum Parsimony (MP) search on PAUP* version 4.0b10 [46], with the tree from our phylogenetic analysis of extant species used as the backbone constraint.
Dating estimates
We estimated the divergence times within Carya using the combined molecular data set in BEAST version 1.7.4 [47]. BEAST employs a Bayesian MCMC to co-estimate topology, substitution rates, and node ages. Based on our phylogenetic analyses of extant and extinct genera in the Juglandaceae, four fossils were used for calibration purposes. Two fossils of extinct genera were used as minimum-age calibration points. Paleooreomunneastoneana is from the middle Eocene [48], and it is sister to the Oreomunnea - Alfaroa clade; we therefore used 40.4 to 48.6 Ma to calibrate the node age of the Oreomunnea - Alfaroa -Engelhardia clade [49]. Polypteramanningii is from the early Paleocene [50] and is sister to subtribe Juglandinae (including Cyclocarya , Juglans , and Pterocarya ) and Platycarya ; and we used its minimum age 64 Ma [49] to constrain the stem of the Juglandinae clade. Additionally, Cyclocarya brownii is an extinct species of Cyclocarya , which now comprises a single living species Cyclocarya paliurus in China [51]; thus we used 55.8 to 61.6 Ma [49] to constrain the stem age of Cyclocarya . These three calibration points were constrained with the lognormal prior distribution. In each implementation, the minimum age of each fossil was used as an offset value. In addition, we adjusted standard deviation and the mean in log space to match the maximum bound of each fossil with the 97.5% of the distribution [52]. An age of 84 Ma was used to constrain the maximum root age of the phylogeny, which is the age of the earliest Fagales fossil Antiquacupula [53-55], with a normal prior distribution (standard deviation, 0.5) [49,55].
The data set was partitioned based on plastid or nuclear genomes. According to the results of jModelTest version 1.0, we used the GTR + Г model as described above. A starting tree with branch lengths satisfying all fossil prior constraints was created using the program r8s version 1.7 [56] by non-parametric rate smoothing (NPRS) [57]. Beast analysis was performed with an uncorrelated lognormal relaxed molecular clock model for 100,000,000 generations. Adequate sampling and convergence of the chain to stationarity were confirmed by inspection of MCMC samples using Tracer ver. 1.5 [58], as all parameters of the effective sample size (ESS) values were greater than 200, suggesting sufficient sampling in our BEAST analysis. A maximum-credibility tree, representing the maximum a posteriori topology, was calculated after removing 50% burn-in with TreeAnnotator version 1.5.4 [59], which also calculated the mean ages and 95% highest posterior density (95% HPD) intervals for each node.
Biogeographic analyses
After estimating the phylogenetic relationships including fossils, we derived a summary tree at the generic level. Subsequently, according to the records of fruit fossils of Carya (Table S4) [30], we mapped three branches to represent the distributions of Carya in the Eocene, Oligocene, and early Miocene, respectively. Each branch was inserted manually in the newick chronogram along the stem lineage of Carya, with placement according to their position in the geologic time scale. Using this phylogeny, we employed the dispersal-vicariance analysis (DIVA) [60] and a maximum likelihood based method LAGRANGE [61,62] to construct the biogeographic history of Carya. Biogeographic analyses were also conducted without fossil information. Based on the geographical distribution patterns of extant and extinct genera of Juglandaceae (Table S5), we categorized the distributions into the following endemic areas: North America (A), Europe (B), and Asia (C).
Diversification rate analyses
To visualize the temporal diversification in Carya, we produced lineage-through-time (LTT) plots using the R package APE [63]. Plots were produced for 1000 sampled trees from the converged BEAST trees for entire Carya genus, then its EA lineages, and its ENA lineages, respectively. The net diversification rate (r) for each clade within Carya was also estimated following equation (7) of Magallón and Sanderson [64] for crown groups under the assumptions of either no extinction (ε = 0) or high relative extinction (ε = 0.9) to evaluate relative species diversification between clades. Calculations were done using the R package LASER [65] and the crown age of each clade was inferred from the BEAST analysis (Table 1).
Table 1. Net diversification rates (r) estimated for Carya based on the equation (7) of Magallón and Sanderson [59].
| Clade | Crown group age (95% HPD) Ma | Number of species | r (ε = 0) | r (ε = 0.9) |
|---|---|---|---|---|
| Carya | 21.58 (11.07-35.51) | 17 | 0.0991 | 0.0418 |
| ENA Carya | 10.10 (5.26-16.78) | 12 | 0.1774 | 0.0682 |
| EA Carya | 12.81 (5.60-22.04) | 5 | 0.0716 | 0.0218 |
Note: ε, the relative extinction rate
Results
Phylogenetic analyses
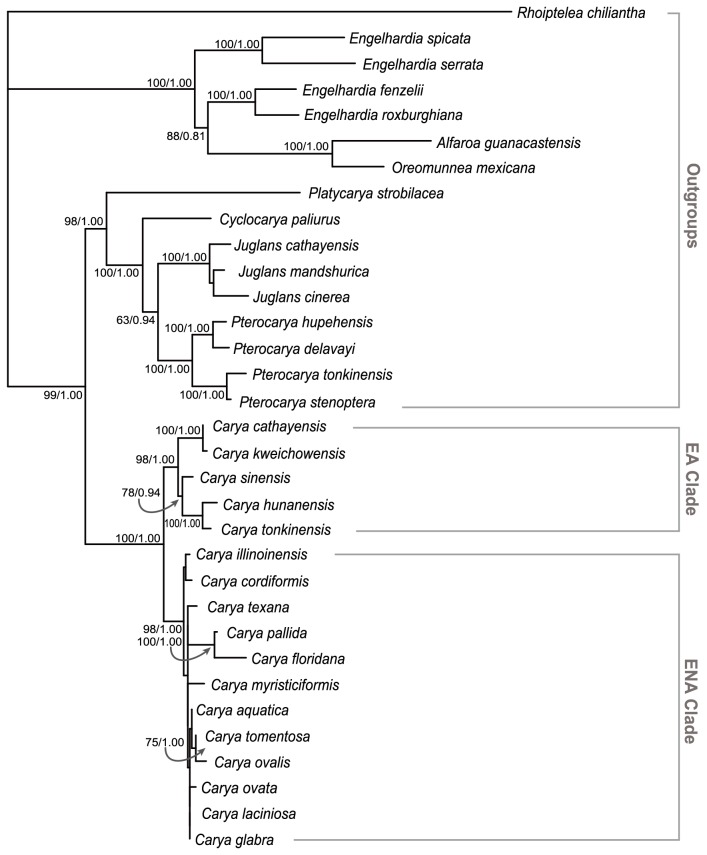
A total of 200 sequences were newly obtained (Table S1). ML and BI result in almost the same trees (Figure 2). Within the Juglandaceae, two major clades are identified. One includes Alfaroa , Engelhardia, and Oreomunnea (maximum likelihood bootstrap support, MLBS 100%; PP 1.00). Carya, Cyclocarya , Juglans , Platycarya , and Pterocarya form the other clade (MLBS 99%; PP 1.00), in which Carya is sister to the clade of the remaining genera.
Figure 2. Phylogeny of Carya based on the combined molecular data set.
ML bootstrap support > 75% and Bayesian posterior probability > 0.80 are indicated at the nodes, respectively. The topologies of strict consensus trees from maximum likelihood and Bayesian inference are same.
Carya is strongly supported as monophyletic (MLBS 100%; PP 1.00), and two major clades are well supported, which correspond to an EA group (MLBS 98%; PP 1.00) and an ENA group (MLBS 98%; PP 1.00), respectively. Within the EA group, C . cathayensis and C . kweichowensis are in a clade that is sister to a subclade of the remaining three species. In the ENA group, the relationships among ENA species are poorly resolved; nevertheless C . ovalis and C . tomentosa form a clade with moderate or strong support (MLBS 75%; PP 1.00).
The placements of extinct genera are resolved in our analysis (Figures 3A and 4A). Three extinct genera, Paleooreomunnea, Palaeocarya, and Paraengelhardia form a clade, sister to the extant Alfaroa - Oreomunnea clade. The extinct Paleoplatycarya and the extant Platycarya form a clade, which is sister to the extinct Hooleya. The extinct Cruciptera is sister to the clade containing extant Juglans and Pterocarya . The extinct Polyptera and the extant Carya are in a clade.
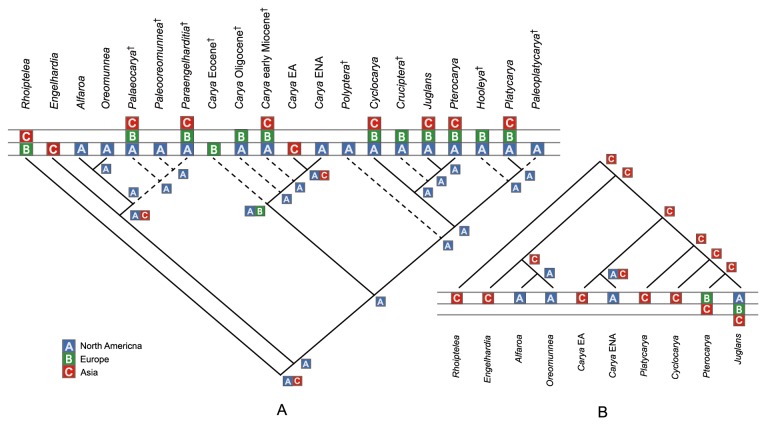
Figure 3. Ancestral area reconstructions for Carya using DIVA.
A: Inference with fossil taxa; B: Inference without fossil taxa. Fossil taxa are indicated by cross symbols after their names. The areas of endemism are defined for both analyses: A, North America; B, Europe; C, Asia.
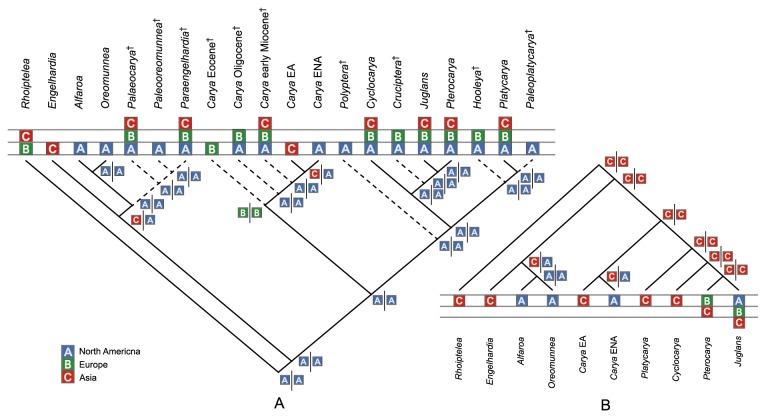
Figure 4. Ancestral area reconstructions for Carya using LAGRANGE.
A: Inference with fossil taxa; B: Inference without fossil taxa. Fossil taxa are indicated by cross symbols after their names. A slash in the results of LAGRANGE indicates the split of areas in two daughter lineages. The areas of endemism are defined for both analyses: A, North America; B, Europe; C, Asia.
Historical biogeography and divergence times
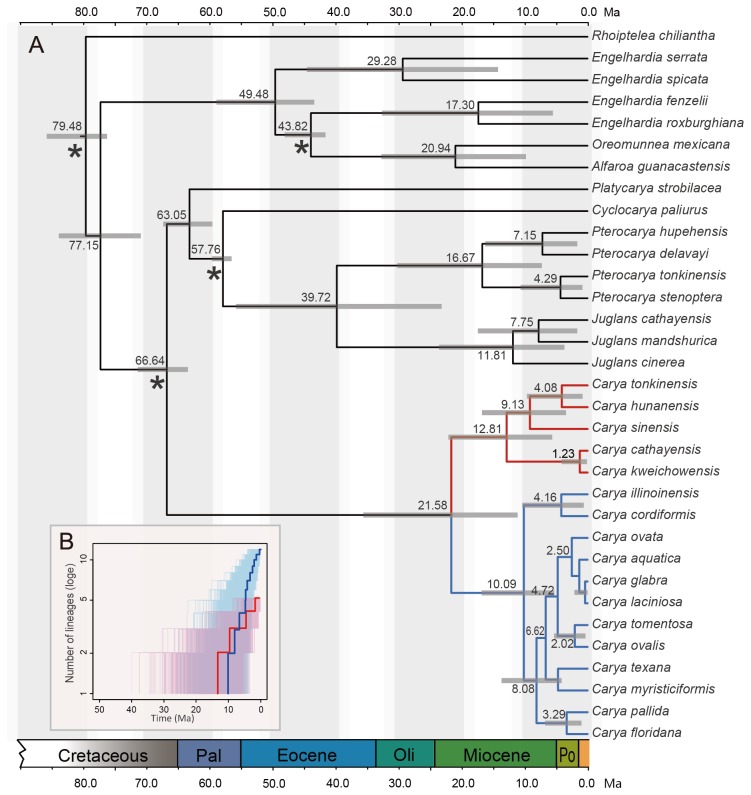
The chronogram of Carya is shown in Figure 5. Based on our dating estimates, the age of the Juglandaceae is 77.15 (95% HPD 70.76-83.76) Ma, and the time of origin of Carya is 66.64 (95% HPD 63.30-71.26) Ma. The split between the EA and ENA clades occurred at 21.58 (95% HPD 11.07-35.51) Ma. EA and ENA Carya began to diversify at 12.81 (95% HPD 5.60-22.04) Ma and 10.10 (95% HPD 5.26-16.78) Ma, respectively.
Figure 5. BEAST chronogram and LTT plots of Carya.
A: The chronogram of Carya inferred from the combined molecular data set. Gray bars represent the 95% HPD intervals of node ages. Calibration points are indicated with asterisks. B: Lineage-through-time (LTT) plots for Carya. The thin lines indicate the 1000 random trees from the best-fitting model obtained in BEAST (EA clade: pink; ENA clade: light blue); the bold lines correspond to the maximum credibility tree from the BEAST analysis (EA clade: red; ENA clade: blue).
Based on the phylogenetic tree including both extant and extinct genera of the Juglandaceae, the DIVA and LAGRANGE analyses suggest that Carya originated in North America (Figures 3A and 4A). In contrast, results from the analyses for the only extant genera of Juglandaceae, however, indicate that the ancestral area of Carya is Asia (Figures 3B and 4B).
Diversification rates
The lineage-through-time (LTT) plots for the EA and ENA clades are shown in Figure 5. Compared with the EA clade, the ENA clade had a dramatic accumulation of lineages after the middle Miocene. Our diversification rate estimates based on net diversification rates (r) following equation (7) of Magallón and Sanderson [64] also showed that the ENA clade had a significantly higher net diversification rate (r) than the EA clade after the middle Miocene (ε = 0, 0.0863 > 0.0389; ε = 0.9, 0.0682 > 0.0218; Table 1).
Discussion
Phylogeny
Our phylogenetic analyses indicate that the Juglandaceae comprise two major clades. Carya is sister to the Cyclocarya - Juglans - Platycarya - Pterocarya clade. The results are congruent with those of previous studies [23,24]. Within Carya, two monophyletic groups corresponding to geographic distributions (EA and ENA) are well supported (EA: MLBS 98%; PP 1.00; ENA: MLBS 98%; PP 1.00). Given that incomplete taxon sampling can sometimes influence phylogenetic inference [66], our analyses with an extensive sampling further clarified the correlation of phylogenetic relationships and geographic distributions in Carya. Although Stone [20] treated C . ovalis as a synonym of C . glabra , our analyses indicate that C . ovalis is sister to C . tomentosa . Morphologically, C . ovalis differs from C . glabra in many characters, such as reddish petioles (vs. plain), fruit stipe rarely present (vs. often present), dehiscence of the fruits to the base (vs. only at apex or to middle), mature husk warty and dull (vs. smooth and shining), and nut shell ridged and thin (vs. not ridged and thick) [22].
Seven extinct genera were placed into the phylogenetic framework of extant species using the DNA scaffold method. Four of these, i.e., Cruciptera, Paleoplatycarya, Paleooreomunnea, and Polyptera, were also used by Manos et al. [24]. The placement of Cruciptera was congruent with that in previous studies [23,24,30]. Paleooreomunnea and the two newly added extinct genera Palaeocarya and Paraengelhardia formed a clade showing a close relationship with an extant clade that includes Alfaroa , Engelhardia, and Oreomunnea . The sister relationship between Paleoplatycarya and Platycarya from our inferences was also recognized in the study of Manos et al. [24]. For the extinct genus Polyptera, previous studies suggested it sister to the clade containing Juglandinae ( Cyclocarya , Juglans , and Pterocarya ) and Carya [50,67]. Our results support Polyptera with a closer relationship to Juglandinae than Carya. This may be ascribed to the similarity of some morphological characters. For example, both Polyptera and Cyclocarya have the pollen with equatorial pores readily distinguished from the subequatorial pores of Carya [23]; the production of a large number of fruits on a spicate infructescence in Polyptera is similar to the situation in Cyclocarya and Pterocarya ; and both Polyptera and Cyclocarya have a disk-like wing oriented perpendicular to the nut axis [50].
Evolution of intercontinental disjunctions in Carya
Both DIVA and LAGRANGE analyses including fossil information suggested North America as the ancestral area for Carya, but analyses without fossil information indicated that Carya originated in Asia. Because Carya has a pollen type known as Caryapollenites, which was suggested to be derived directly from Normapolles [30,68], the origin of Carya should be within or at least near the distribution of the Normapolles complex, which defines a middle to late Cretaceous province including ENA and Europe. Therefore, the palynological evidence supports the inference by the analyses with fossil information for an origin in North America. Although Carya is not native in Europe today, according to the fossil evidence, Europe was a center of diversity of Carya during the Tertiary, especially during the Miocene, with some species extending even to the Quaternary [30]. The difference in ancestral area reconstruction is likely due to the much broader geographic distributions of the fossils. Hence, the fossil information is particularly important for reliably reconstructing the historical biogeography of Carya and the following discussion is based on the result inferred by the analyses that included occurrence data from the fossil records.
Our result for the age of the Juglandaceae, as 77.15 (95% HPD 70.76-83.76) Ma, is similar with the outcome of Sauquet et al. [49] inferred with the default calibrations, as 72.6 (95% HPD 64.4-81.3) Ma. Our estimations are also congruent with those of Manos et al. [24] who constrained the minimum age of the Juglandinae clade at 58 Ma. The age of the subfamily Juglandoideae (including Platycarya , Carya, Cyclocarya , Juglans , and Pterocarya ) is herein estimated as 66.64 [95% HPD 63.30-71.26] Ma, which is almost the same as the estimation of 65.0 [95% HPD 58.3-73.0] Ma by Sauquet et al. [49] using the default calibrations. In addition, based on the presence of fossil pollen which is similar to that of extant Carya of western Europe and North America in the Late Paleocene. Manchester [30] noted that the first hint of the Carinae (the subtribe including Carya) occurred in the Paleocene. This also supports our estimate for the stem age of Carya dating back to the Paleocene (66.64 [95% HPD 63.30-71.26]). Therefore, the divergence date estimates inferred by our calibration points should be reliable to reconstruct the biogeographic history of Carya.
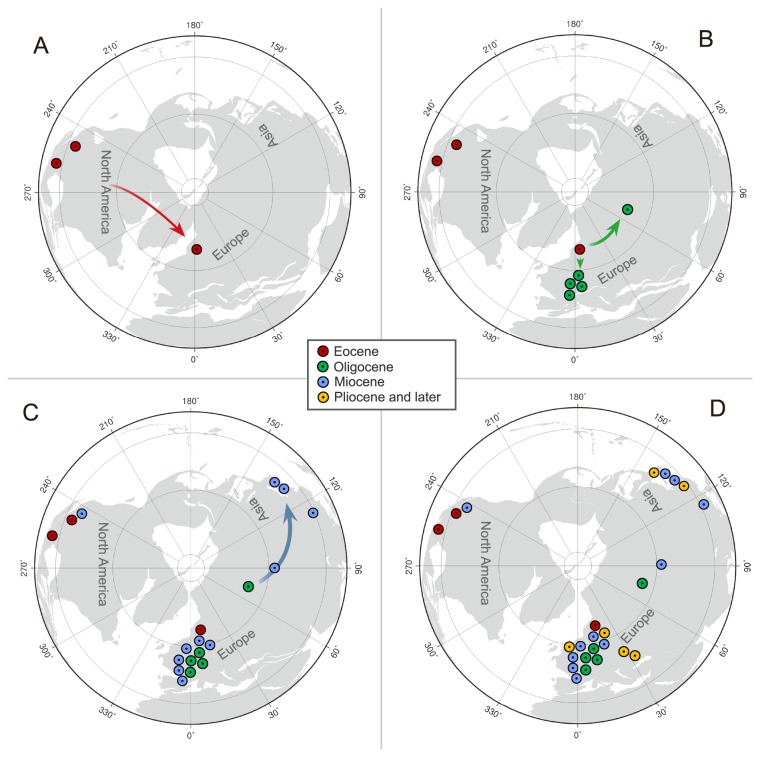
The age estimates and biogeographic analyses suggest Carya originated in North America during the early Paleocene. Previous studies have inferred that a relatively uniform, warm climate was prevalent in the Northern Hemisphere during the early Tertiary, which allowed populations of the boreotropical flora to move between continental areas via northern land bridges [4-6,15,34]. Both the BLB and NALB played important roles in plant exchanges during the Tertiary, but with differential importance in different geologic times [34]. Previous studies suggest that the floristic migration via the NALB was possible during the Paleocene and Eocene, and it was more likely to be used by thermophilic species [5,6,13,15]. The BLB was used mostly by temperate taxa during the late Miocene and Pliocene (<10 Ma [4,8,14]). The divergence time of the two Carya clades indicates that Carya likely migrated from North America to Europe via the NALB like other thermophilic groups, such as Ampelopsis (Vitaceae) [69], Cercis (Fabaceae) [70], Cornus (Cornaceae) [35], Malpighiaceae [71], and Quercus (Fagaceae) [72]. The distribution pattern of Carya fossils also supports this inference. Given that the NALB connected North America and western Europe, Carya-like pollen was abundant in both continents and nuts have been confirmed from the late Eocene of North America [31]. Oligocene and younger macrofossils of Carya have been confirmed in western Europe, supporting an Oligocene regional diversification. In contrast, the earliest eastern Asian Carya fruit fossils occurred only in the Miocene (Figure 6). The group might have entered Asia from Europe as the Turgai seaway receded, and/or from North America via the BLB.
Figure 6. Hypothetical migratory routes of Carya.
A: Eocene; B: Oligocene; C: Miocene; D: Pliocene and later. The colored arrows and circles indicate the migratory directions and Carya fruit fossil records, respectively.
Starting in the Miocene, there was a distinct climatic cooling period across the high latitude areas of the Northern Hemisphere, which may have resulted in a reduction of the distribution of forests [5]. According to the maps of the past geographic ranges of Carya presented by Manchester [30] and Mai [32], Carya was more broadly distributed in the Miocene, with occurrences including Europe, Siberia, and western North America, where the genus is absent today. The present disjunct distribution of Carya between EA and ENA can be explained by extinctions in large parts of its former ranges. Therefore, extinctions caused the range fragmentation of Carya, ultimately leading to the modern intercontinental distribution between EA and ENA. Extinction events could have extirpated the old stem relatives that diverged prior to the extant crown radiation, leaving a phylogeny that includes only extant taxa with long stems and species-rich crowns [73]. A remarkably long “temporal gap” between the Carya stem age (the Early Cretaceous) and the beginning of the extant radiation (the Early Miocene) is detected, which supports that Carya has been experienced several extinction events during the Tertiary.
Higher diversification rate in the ENA Clade
Our results of diversification analyses showed that ENA Carya has a significantly higher diversification rate than EA since the Miocene. Both environmental [74-77] and biological attributes [78-80] may directly facilitate species diversification [81-83]. The formation of geologic barriers is one of the most important factors driving speciation [84-87]. For ENA, after being largely eroded to plains by the end of the Mesozoic, renewed uplift of Appalachian Mountains occurred during the Tertiary [88,89]. According to the data from the sedimentation rates and fault ages, this major uplift occurred during the late Oligocene to Miocene, followed by a quiescent interval, in which erosion occurred that lasted until the end of Miocene [90-96]. These dramatic changes of environments offered many ecological opportunities, contributing to divergence in several taxa in this area [87,97-103]. This is consistent with our inference that ENA Carya underwent a high diversification rate since the late Miocene. On the other hand, EA Carya taxa are primarily distributed in central, south-central, and south-eastern China, a region that was relatively stable tectonically since the late Tertiary [104,105]. This stability allowed these areas to be refugia, facilitating the survival of many relict plant lineages during the cold period of the Tertiary. Once climatic conditions improved, not all refugial species were able to migrate out but a significant proportion of species remained restricted [105-108]. Hence, these species exhibit disjunct distributions and relatively lower diversification rates [109]. The EA Carya lineage is a typical example of this pattern. Although during [110-112] or immediately before [113,114] the Pliocene and Pleistocene, the major uplift of the Tibetan Plateau took place in EA, which created a vast array of new habitats across wide elevational ranges in the eastern fringe of the Tibetan Plateau [115], the distribution areas of EA Carya are not part of this region.
ENA Carya species have terminal bud scales, while the EA species have naked terminal buds (excluding C. sinensis with pseudo-valvate terminal bud scales). The presence of bud scales is considered to be an adaptation for cooling climates [116], so the ENA species may be better adapted for the cooler temperate climate since the Miocene. At least six ENA Carya species ( C . floridana , C . glabra , C. myristiciformis , C . pallida , C . texana , and C . tomentosa ) are tetraploid (n = 32) [20,117,118], and the only confirmed count for EA Carya species is diploid ( C . cathayensis , n = 16 [20,119]). Polyploidization may allow species to adapt to dramatic changes in the environment [118,120,121], so that polyploid lineages may have higher diversification rates than that of diploid lineages [122,123]. Furthermore, sympatric Carya species in southeastern Ohio have shown distribution replacement especially concerning topography and its associated microhabitats [124]. Thus, the richness of ecological opportunities, key morphological innovations, and polyploidy may have been responsible for the high diversification rate of ENA Carya.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
The aligned sequence data is presented in Nexus format.
(TXT)
Acknowledgments
The authors thank Douglas E. Soltis and Pamela S. Soltis for carefully reading an early version of the manuscript. We also would like to thank Jun-Xia Su, Hong-Lei Li, Min Chen, Miao Sun, Tuo Yang, and Xiao-Yu Dong for laboratory assistance; Chih-Chieh Yu and Li-Min Lu for providing helpful advice to improve an early version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (NNSF 31270268 and NNSF 31061160184) and Chinese Academy of Sciences Visiting Professorship for Senior International Scientists (2011T1S24). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wen J (1999) Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Annu Rev Ecol Syst 30: 421-455. doi:10.1146/annurev.ecolsys.30.1.421. [Google Scholar]
- 2. Manos PS, Donoghue MJ (2001) Progress in Northern Hemisphere phytogeography: an introduction. Int J Plant Sci 162: S1-S2. doi:10.1086/317898. [Google Scholar]
- 3. Wen J (2001) Evolution of eastern Asian-eastern North American biogeographic disjunctions: a few additional issues. Int J Plant Sci 162: 117-122. doi:10.1086/322940. [Google Scholar]
- 4. Wang W, Chen ZD, Liu Y, Li RQ, Li JH (2007) Phylogenetic and biogeographic diversification of Berberidaceae in the northern hemisphere. Syst Bot 32: 731-742. doi:10.1600/036364407783390791. [Google Scholar]
- 5. Wolfe JA (1975) Some aspects of plant geography of the Northern Hemisphere during the Late Cretaceous and Tertiary. Ann Mo Bot Gard 62: 264-279. doi:10.2307/2395198. [Google Scholar]
- 6. Tiffney BH (1985) Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J Arnold Arboretum 66: 73-94. [Google Scholar]
- 7. Taylor DW (1990) Paleobiogeographic relationships of angiosperms from the Cretaceous and early Tertiary of the North American area. Bot Rev 56: 279-417. doi:10.1007/BF02995927. [Google Scholar]
- 8. Xiang QY, Soltis DE, Soltis PS, Manchester SR, Crawford DJ (2000) Timing the eastern Asian-eastern North American floristic disjunction: molecular clock corroborates paleontological estimates. Mol Phylogenet Evol 15: 462-472. doi:10.1006/mpev.2000.0766. PubMed: 10860654. [DOI] [PubMed] [Google Scholar]
- 9. Donoghue MJ, Smith SA (2004) Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos Trans R Soc Lond B Biol Sci 359: 1633-1644. doi:10.1098/rstb.2004.1538. PubMed: 15519978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hopkins D (1967) The Bering land bridge. Palo Alto: Stanford University Press. [Google Scholar]
- 11. Budantsev LY (1992) Early stages of formation and dispersal of the temperate flora in the boreal region. Bot Rev 58: 1-48. doi:10.1007/BF02858542. [Google Scholar]
- 12. McKenna MC (1983) Holarctic landmass rearrangement, cosmic events, and Cenozoic terrestrial organisms. Ann Mo Bot Gard 70: 459-489. doi:10.2307/2992083. [Google Scholar]
- 13. Tiffney BH (1985) The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J Arnold Arboretum 66: 243-273. [Google Scholar]
- 14. Wen J, Ickert-Bond S, Nie Z, Li R (2010) Timing and modes of evolution of eastern Asian–North American biogeographic disjunctions in seed plants. In: Long M, Gu H, Zhou Z. Darwin’s Heritage Today : Proceedings of the Darwin 200 Beijing International Conference. Beijing: Higher Education Press. [Google Scholar]
- 15. Tiffney BH, Manchester SR (2001) The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. Int J Plant Sci 162: S3-S17. doi:10.1086/323880. [Google Scholar]
- 16. Donoghue MJ, Bell CD, Li J (2001) Phylogenetic patterns in Northern Hemisphere plant geography. Int J Plant Sci 162: S41-S52. doi:10.1086/323278. [Google Scholar]
- 17. Wu ZY (2003) The Families and Genera of Angiosperms in China: a comprehensive analysis. Beijing: Science Press. [Google Scholar]
- 18. Hong D-Y (1993) Eastern Asian–North American disjunctions and their biological significance. Cathaya 5: 1-39. [Google Scholar]
- 19. Xiang QYJ, Zhang WH, Ricklefs RE, Qian H, Chen ZD et al. (2004) Regional differences in rates of plant speciation and molecularevolution: a comparison between eastern Asia and eastern North America. Evolution 58: 2175-2184. doi:10.1111/j.0014-3820.2004.tb01596.x. PubMed: 15568220. [DOI] [PubMed] [Google Scholar]
- 20. Stone DE (1997) Juglandaceae. In: Flora of North America Editorial Committee editors. Flora of North America north of Mexico. New York: Oxford University Press. [Google Scholar]
- 21. Lu AM, Stone DE, Grauke LJ (1999) Juglandaceae. In: Wu ZY, Hong DY, Raven PH. Flora of China. Beijing and Missouri: Science Press and Missouri Botanical Garden Press. [Google Scholar]
- 22. Manning WE (1950) A key to the Hickories north of Virginia with notes on the two Pignuts Carya glabra and C. ovalis. Rhodora 52: 188-199. [Google Scholar]
- 23. Manos PS, Stone DE (2001) Evolution, phylogeny, and systematics of the Juglandaceae. Ann Mo Bot Gard 88: 231-269. doi:10.2307/2666226. [Google Scholar]
- 24. Manos PS, Soltis PS, Soltis DE, Manchester SR, Oh SH et al. (2007) Phylogeny of extant and fossil Juglandaceae inferred from the integration of molecular and morphological data sets. Syst Biol 56: 412-430. doi:10.1080/10635150701408523. PubMed: 17558964. [DOI] [PubMed] [Google Scholar]
- 25. Leroy JF (1955) Étude sur les Juglandaceae. à la recherche d'une conception morphologique de la fleur femelle et du fruit. Bulletin Museums Natl Histoire Nat 6B: 1-246. [Google Scholar]
- 26. Manning WE (1963) Hickories reported in India and Laos with other notes on Carya in Asia. Brittonia 15: 123-125. doi:10.2307/2805397. [Google Scholar]
- 27. Pollock DD, Zwickl DJ, McGuire JA, Hillis DM (2002) Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol 51: 664-671. doi:10.1080/10635150290102357. PubMed: 12228008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zwickl DJ, Hillis DM (2002) Increased taxon sampling greatly reduces phylogenetic error. Syst Biol 51: 588-598. doi:10.1080/10635150290102339. PubMed: 12228001. [DOI] [PubMed] [Google Scholar]
- 29. Müller MJ (1982) Selected climatic data for a global set of standard stations for vegetation science. Hague: Dr. W. Junk Publishers. [Google Scholar]
- 30. Manchester SR (1987) The fossil history of the Juglandaceae. Monogr Syst Bot From Mo Bot Gard 21: 1-137. [Google Scholar]
- 31. Manchester SR, McIntosh WC (2007) Late Eocene silicified fruits and seeds from the John Day Formation near Post, Oregon. Paleobios 27: 7-17. [Google Scholar]
- 32. Mai DH (1981) Der Formenkreis der Vietnam-Nuß (Carya poilanei (Chev.) Leroy) in Europa. Feddes Repertorium 92: 339-385. [Google Scholar]
- 33. Mai DH (1995) Tertiäre Vegetationsgeschichte Europas: Methoden und Ergebnisse. Heidelberg: Spektrum Akademischer Verlag. [Google Scholar]
- 34. Xiang QYJ, Manchester SR, Thomas DT, Zhang W, Fan C (2005) Phylogeny, biogeography, and molecular dating of cornelian cherries (Cornus, Cornaceae): tracking Tertiary plant migration. Evolution 59: 1685-1700. doi:10.1554/03-763.1. PubMed: 16329240. [PubMed] [Google Scholar]
- 35. Xiang QYJ, Thomas DT, Zhang W, Manchester SR, Murrell Z (2006) Species level phylogeny of the genus Cornus (Cornaceae) based on molecular and morphological evidenceimplications for taxonomy and Tertiary intercontinental migration. Taxon 55: 9-30. doi:10.2307/25065525. [Google Scholar]
- 36. Hillis DM (1987) Molecular versus morphological approaches to systematics. Annu Rev Ecol Syst 18: 23-42. doi:10.1146/annurev.es.18.110187.000323. [Google Scholar]
- 37. Springer MS, Teeling EC, Madsen O, Stanhope MJ, De Jong WW (2001) Integrated fossil and molecular data reconstruct bat echolocation. Proc Natl Acad Sci USA 98: 6241-6246. doi:10.1073/pnas.111551998. PubMed: 11353869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manchester SR (1991) Cruciptera, a new juglandaceous winged fruit from the Eocene and Oligocene of western North America. Syst Bot 16: 715-725. doi:10.2307/2418873. [Google Scholar]
- 39. Li RQ, Chen ZD, Lu AM, Soltis DE, Soltis PS et al. (2004) Phylogenetic relationships in Fagales based on DNA sequences from three genomes. Int J Plant Sci 165: 311-324. doi:10.1086/381920. [Google Scholar]
- 40. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. doi:10.1093/bioinformatics/btm404. PubMed: 17846036. [DOI] [PubMed] [Google Scholar]
- 41. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/ 98/ NT. Nucleic Acids Symp Series 41: 95-98. [Google Scholar]
- 42. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688-2690. doi:10.1093/bioinformatics/btl446. PubMed: 16928733. [DOI] [PubMed] [Google Scholar]
- 43. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758-771. doi:10.1080/10635150802429642. PubMed: 18853362. [DOI] [PubMed] [Google Scholar]
- 44. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253-1256. doi:10.1093/molbev/msn083. PubMed: 18397919. [DOI] [PubMed] [Google Scholar]
- 45. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. doi:10.1093/bioinformatics/btg180. PubMed: 12912839. [DOI] [PubMed] [Google Scholar]
- 46. Swofford DL (2003) PAUP*: Phylogenetic analysis using parsimony (and other methods), version 4. Sunderland (Massachusetts): Sinauer Associates. [Google Scholar]
- 47. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. doi:10.1186/1471-2148-7-214. PubMed: 17996036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dilcher DL, Potter FW Jr, Crepet WL (1976) Investigations of angiosperms from the Eocene of North America: Juglandaceous winged fruits. Am J Bot 63: 532-544. doi:10.2307/2441817. [Google Scholar]
- 49. Sauquet H, Ho SYW, Gandolfo MA, Jordan GJ, Wilf P et al. (2012) Testing the Impact of Calibration on Molecular Divergence Times Using a Fossil-Rich Group: The Case of Nothofagus (Fagales). Syst Biol 61: 289-313. doi:10.1093/sysbio/syr116. PubMed: 22201158. [DOI] [PubMed] [Google Scholar]
- 50. Manchester SR, Dilcher DL (1997) Reproductive and vegetative morphology of Polyptera (Juglandaceae) from the Paleocene of Wyoming and Montana, American Journal of Botany 84: 649-649 [PubMed]
- 51. Wing SL, Hickey LJ (1984) The Platycarya perplex and the evolution of the Juglandaceae. Am J Bot 71: 388-411. doi:10.2307/2443497. [Google Scholar]
- 52. Warnock RC, Yang Z, Donoghue PC (2012) Exploring uncertainty in the calibration of the molecular clock. Biol Lett 8: 156-159. doi:10.1098/rsbl.2011.0710. PubMed: 21865245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proc R Soc Lond B Biol Sci 268: 2211-2220. doi:10.1098/rspb.2001.1782. PubMed: 11674868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sims HJ, Herendeen PS, Crane PR (1998) New genus of fossil Fagaceae from the Santonian (Late Cretaceous) of central Georgia, USA. Int J Plant Sci 159: 391-404. doi:10.1086/297559. [Google Scholar]
- 55. Ho SY, Phillips MJ (2009) Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol 58: 367-380. doi:10.1093/sysbio/syp035. PubMed: 20525591. [DOI] [PubMed] [Google Scholar]
- 56. Sanderson MJ (2003) r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19: 301-302. doi:10.1093/bioinformatics/19.2.301. PubMed: 12538260. [DOI] [PubMed] [Google Scholar]
- 57. Sanderson MJ (1997) A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol 14: 1218-1231. doi:10.1093/oxfordjournals.molbev.a025731. [Google Scholar]
- 58. Rambaut A, Drummond A (2007) Tracer v1. 5. Available: http://tree.bio.ed.ac.uk/software/tracer.
- 59. Rambaut A, Drummond A (2007) A guide to TreeAnnotator, version 1.5.4. Available: http://beast.bio.ed.ac.uk/MainPage.
- 60. Ronquist F (1997) Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol 46: 195-203. doi:10.1093/sysbio/46.1.195. [Google Scholar]
- 61. Ree RH, Moore BR, Webb CO, Donoghue MJ (2005) A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59: 2299-2311. doi:10.1111/j.0014-3820.2005.tb00940.x. PubMed: 16396171. [PubMed] [Google Scholar]
- 62. Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol 57: 4-14. doi:10.1080/10635150701883881. PubMed: 18253896. [DOI] [PubMed] [Google Scholar]
- 63. Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289-290. doi:10.1093/bioinformatics/btg412. PubMed: 14734327. [DOI] [PubMed] [Google Scholar]
- 64. Magallón S, Sanderson MJ (2001) Absolute diversification rates in angiosperm clades. Evolution 55: 1762-1780. doi:10.1111/j.0014-3820.2001.tb00826.x. PubMed: 11681732. [DOI] [PubMed] [Google Scholar]
- 65. Rabosky DL (2006) LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinform Online 2: 247 PubMed: 19455217. [PMC free article] [PubMed] [Google Scholar]
- 66. Nabhan AR, Sarkar IN (2011) The impact of taxon sampling on phylogenetic inference: a review of two decades of controversy. Brief Bioinform 13: 1-13. PubMed: 21441561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Manchester SR, Dilcher DL (1982) Pterocaryoid fruits (Juglandaceae) in the Paleogene of North America and their evolutionary and biogeographic significance. Am J Bot 69: 275-286. doi:10.2307/2443015. [Google Scholar]
- 68. Traverse A (2007) Paleopalynology. Netherlands: Kluwer Academic Pub.
- 69. Nie ZL, Sun H, Manchester SR, Meng Y, Luke Q et al. (2012) Evolution of the intercontinental disjunctions in six continents in the Ampelopsis clade of the grape family (Vitaceae). BMC Evol Biol 12: 17-30. doi:10.1186/1471-2148-12-17. PubMed: 22316163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Davis CC, Fritsch PW, Li J, Donoghue MJ (2002) Phylogeny and biogeography of Cercis (Fabaceae): evidence from nuclear ribosomal ITS and chloroplast ndhF sequence data. Syst Bot 27: 289-302. [Google Scholar]
- 71. Davis CC, Bell CD, Fritsch PW, Mathews S (2002) Phylogeny of Acridocarpus-Brachylophon (Malpighiaceae): implications for Tertiary tropical floras and Afroasian biogeography. Evolution 56: 2395-2405. doi:10.1554/0014-3820(2002)056[2395:POABMI]2.0.CO;2. PubMed: 12583580. [DOI] [PubMed] [Google Scholar]
- 72. Denk T, Grímsson F, Zetter R (2010) Episodic migration of oaks to Iceland: Evidence for a North Atlantic “land bridge” in the latest Miocene. Am J Bot 97: 276-287. doi:10.3732/ajb.0900195. PubMed: 21622388. [DOI] [PubMed] [Google Scholar]
- 73. Antonelli A, Sanmartín I (2011) Mass extinction, gradual cooling, or rapid radiation? Reconstructing the spatiotemporal evolution of the ancient angiosperm genus Hedyosmum (Chloranthaceae) using empirical and simulated approaches. Syst Biol 60: 596-615. doi:10.1093/sysbio/syr062. PubMed: 21856636. [DOI] [PubMed] [Google Scholar]
- 74. Cracraft J (1985) Biological diversification and its causes. Ann Mo Bot Gard 72: 794-822. doi:10.2307/2399222. [Google Scholar]
- 75. Ricklefs RE, Schluter D (1993) Species diversity in ecological communities: historical and geographical perspectives. Chicage. University of Chicago Press. [Google Scholar]
- 76. Guo Q, Ricklefs RE, Cody ML (1998) Vascular plant diversity in eastern Asia and North America: historical and ecological explanations. Bot J Linn Soc 128: 123-136. doi:10.1111/j.1095-8339.1998.tb02111.x. [Google Scholar]
- 77. Givnish TJ, Barfuss MHJ, Van Ee B, Riina R, Schulte K et al. (2011) Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Am J Bot 98: 872-895. doi:10.3732/ajb.1000059. PubMed: 21613186. [DOI] [PubMed] [Google Scholar]
- 78. Barraclough TG, Harvey PH, Nee S (1995) Sexual selection and taxonomic diversity in passerine birds. Proc R Soc Lond B Biol Sci 259: 211-215. doi:10.1098/rspb.1995.0031. [Google Scholar]
- 79. Hodges SA, Arnold ML (1995) Spurring plant diversification: are floral nectar spurs a key innovation? Proc R Soc Lond B Biol Sci 262: 343-348. doi:10.1098/rspb.1995.0215. [Google Scholar]
- 80. Mitra S, Landel H, Pruett-Jones S (1996) Species richness covaries with mating system in birds. Auk 113: 544-551. doi:10.2307/4088974. [Google Scholar]
- 81. Rabosky DL, Donnellan SC, Talaba AL, Lovette IJ (2007) Exceptional among-lineage variation in diversification rates during the radiation of Australia’s most diverse vertebrate clade. Proc R Soc Lond B Biol Sci 274: 2915-2923. doi:10.1098/rspb.2007.0924. PubMed: 17878143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rabosky DL (2009) Ecological limits on clade diversification in higher taxa. Am Nat 173: 662-674. doi:10.1086/597378. PubMed: 19302027. [DOI] [PubMed] [Google Scholar]
- 83. Rabosky DL, Adams DC (2012) Rates of morphological evolution are correlated with species richness in salamanders. Evolution 66: 1807–1818. doi:10.1111/j.1558-5646.2011.01557.x. PubMed: 22671548. [DOI] [PubMed] [Google Scholar]
- 84. Mayr E (1942) Systematics and the origin of species, from the viewpoint of a zoologist. London: Harvard University Press. [Google Scholar]
- 85. Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405: 907-913. doi:10.1038/35016000. PubMed: 10879524. [DOI] [PubMed] [Google Scholar]
- 86. Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci 359: 183-195. doi:10.1098/rstb.2003.1388. PubMed: 15101575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lemmon EM, Lemmon AR, Cannatella DC (2007) Geological and climatic forces driving speciation in the continentally distributed trilling chorus frogs (Pseudacris). Evolution 61: 2086-2103. doi:10.1111/j.1558-5646.2007.00181.x. PubMed: 17767584. [DOI] [PubMed] [Google Scholar]
- 88. Dunbar CO, Waagé KM (1949) Historical geology. New York: Wiley. [Google Scholar]
- 89. Cleaves ET (1989) Appalachian Piedmont landscapes from the Permian to the Holocene. Geomorphology 2: 159-179. doi:10.1016/0169-555X(89)90010-X. [Google Scholar]
- 90. Prowell DC, O’Connor BJ (1978) Belair fault zone: Evidence of Tertiary fault displacement in eastern Georgia. Geology 6: 681-684. doi:10.1130/0091-7613(1978)6. [Google Scholar]
- 91. Hack JT (1982) Physiographic divisions and differential uplift in the Piedmont and Blue Ridge. Washington, DC: US Government Printing Office. [Google Scholar]
- 92. Reinhardt J, Prowell DC, Christopher RA (1984) Evidence for Cenozoic tectonism in the southwest Georgia Piedmont. Geol Soc Am Bull 95: 1176-1187. doi:10.1130/0016-7606(1984)95. [Google Scholar]
- 93. Poag CW, Sevon WD (1989) A record of Appalachian denudation in postrift Mesozoic and Cenozoic sedimentary deposits of the US middle Atlantic continental margin. Geomorphology 2: 119-157. doi:10.1016/0169-555X(89)90009-3. [Google Scholar]
- 94. Prowell DC, Christopher R (2000) The last Appalachian orogeny: evidence for Cenozoic tectonism and uplift of mountains in the eastern United States. Geological Society of America, Southeastern Section Meeting. Abstracts with Programs 32: 67. [Google Scholar]
- 95. Prowell DC, Christopher R (2006) Evidence for late Cenozoic uplift in the southern Appalachian Mountains from isolated sediment traps. Geological Society of America, Southeastern Section Meeting. Abstracts with Programs 38: 67. [Google Scholar]
- 96. Dennison JM, Stewart K (2001) Regional structural and stratigraphic evidence for dating Cenozoic uplift of Southern Appalachian highlands. Geological Society of America, Southeastern Section Meeting. Abstracts with Programs 50: 5. [Google Scholar]
- 97. Austin JD, Lougheed SC, Boag PT (2004) Discordant temporal and geographic patterns in maternal lineages of eastern north American frogs, Rana catesbeiana (Ranidae) and Pseudacris crucifer (Hylidae). Mol Phylogenet Evol 32: 799-816. doi:10.1016/j.ympev.2004.03.006. PubMed: 15288057. [DOI] [PubMed] [Google Scholar]
- 98. Austin JD, Lougheed SC, Neidrauer L, Chek AA, Boag PT (2002) Cryptic lineages in a small frog: the post-glacial history of the spring peeper, Pseudacris crucifer (Anura: Hylidae). Mol Phylogenet Evol 25: 316-329. doi:10.1016/S1055-7903(02)00260-9. PubMed: 12414313. [DOI] [PubMed] [Google Scholar]
- 99. Burbrink FT, Lawson R, Slowinski JB (2000) Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): a critique of the subspecies concept. Evolution 54: 2107-2118. doi:10.1554/0014-3820(2000)054[2107:MDPOTP]2.0.CO;2. PubMed: 11209786. [DOI] [PubMed] [Google Scholar]
- 100. Church SA, Kraus JM, Mitchell JC, Church DR, Taylor DR (2003) Evidence for multiple Pleistocene refugia in the postglacial expansion of the eastern tiger salamander, Ambystoma tigrinum . Evolution 57: 372-383. doi:10.1111/j.0014-3820.2003.tb00271.x. PubMed: 12683533. [DOI] [PubMed] [Google Scholar]
- 101. Leaché AD, Reeder TW (2002) Molecular systematics of the eastern fence lizard (Sceloporus undulatus): a comparison of parsimony, likelihood, and Bayesian approaches. Syst Biol 51: 44-68. doi:10.1080/106351502753475871. PubMed: 11943092. [DOI] [PubMed] [Google Scholar]
- 102. Runck AM, Cook JA (2005) Postglacial expansion of the southern red-backed vole (Clethrionomys gapperi) in North America. Mol Ecol 14: 1445-1456. doi:10.1111/j.1365-294X.2005.02501.x. PubMed: 15813783. [DOI] [PubMed] [Google Scholar]
- 103. Zamudio KR, Savage WK (2003) Historical isolation, range expansion, and secondary contact of two highly divergent mitochondrial lineages in spotted salamanders (Ambystoma maculatum). Evolution 57: 1631-1652. doi:10.1554/02-342. PubMed: 12940367. [DOI] [PubMed] [Google Scholar]
- 104. Hsu J (1983) Late Cretaceous and Cenozoic vegetation in China, emphasizing their connections with North America. Ann Mo Bot Gard 70: 490-508. doi:10.2307/2992084. [Google Scholar]
- 105. López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S (2011) Centres of plant endemism in China: places for survival or for speciation? J Biogeogr 38: 1267-1280. doi:10.1111/j.1365-2699.2011.02504.x. [Google Scholar]
- 106. Crisp MD, Laffan S, Linder HP, Monro A (2001) Endemism in the Australian flora. J Biogeogr 28: 183-198. doi:10.1046/j.1365-2699.2001.00524.x. [Google Scholar]
- 107. Linder H (2001) Plant diversity and endemism in sub-Saharan tropical Africa. J Biogeogr 28: 169-182. doi:10.1046/j.1365-2699.2001.00527.x. [Google Scholar]
- 108. Tribsch A (2004) Areas of endemism of vascular plants in the Eastern Alps in relation to Pleistocene glaciation. J Biogeogr 31: 747-760. doi:10.1111/j.1365-2699.2004.01065.x. [Google Scholar]
- 109. IlJinskaya (1970) Monograph of the genus Pterocarya Kunth. Tr Botanicheskogo Instituta Akad Nauk SSSR 10: 7-123. [Google Scholar]
- 110. Li J, Fang X (1999) Uplift of the Tibetan Plateau and environmental changes. Chin Sci Bull 44: 2117-2124. doi:10.1007/BF03182692. [Google Scholar]
- 111. Zhang D, Fengquan L, Jianmin B (2000) Eco-environmental effects of the Qinghai–Tibet Plateau uplift during the Quaternary in China. Environ Geol 39: 1352-1358. doi:10.1007/s002540000174. [Google Scholar]
- 112. Zheng H, Powell CM, An Z, Zhou J, Dong G (2000) Pliocene uplift of the northern Tibetan Plateau. Geology 28: 715-718. doi:10.1130/0091-7613(2000)028. [Google Scholar]
- 113. Harrison TM, Copeland P, Kidd WS, Yin A (1992) Raising tibet. Science 255: 1663-1670. doi:10.1126/science.255.5052.1663. PubMed: 17749419. [DOI] [PubMed] [Google Scholar]
- 114. An ZS, Kutzbach JE, Prell WL, Porter SC (2001) Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature 411: 62-66. doi:10.1038/35075035. PubMed: 11333976. [DOI] [PubMed] [Google Scholar]
- 115. Liu J, Tian B (2007) Origin, evolution, and systematics of Himalaya endemic genera. Newsletter Himalayan Bot 40: 20-27. [Google Scholar]
- 116. Lu AM (1982) On the geographical distribution of the Juglandaceae. Acta Phytotaxonomica Sin 20: 257-274. [Google Scholar]
- 117. Little EL (1971) Conifers and important hardwoods. Atlas of United States trees: Vol. 1. Washington DC: Miscullaneous Publication. [Google Scholar]
- 118. Stone DE (1962) Affinities of a Mexican endemic, Carya palmeri, with American and Asian hickories. Am J Bot 49: 199-212. doi:10.2307/2439542. [Google Scholar]
- 119. Chen R (1993) Chromosome Atlas of Chinese Fruit Trees and Their Close Wild Relatives. Chromosome Atlas of Chinese Principal Economic Plants, Vol. 1 Beijing: Internatinal Academic Publishers. [Google Scholar]
- 120. Fawcett JA, Maere S, Van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc Natl Acad Sci USA 106: 5737–5742. doi:10.1073/pnas.0900906106. PubMed: 19325131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Van de Peer Y, Maere S, Meyer A (2009) The evolutionary significance of ancient genome duplications. Nat Rev Genet 10: 725-732. doi:10.1038/nrg2600. PubMed: 19652647. [DOI] [PubMed] [Google Scholar]
- 122. Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH et al. (2009) Polyploidy and angiosperm diversification. Am J Bot 96: 336-348. doi:10.3732/ajb.0800079. PubMed: 21628192. [DOI] [PubMed] [Google Scholar]
- 123. Soltis DE, Burleigh JG (2009) Surviving the KT mass extinction: New perspectives of polyploidization in angiosperms. Proc Natl Acad Sci USA 106: 5455-5456. doi:10.1073/pnas.0901994106. PubMed: 19336584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. McCarthy BC, Wistendahl WA (1988) Hickory (Carya spp.) distribution and replacement in a second-growth oak hickory forest of southeastern Ohio. American Midland Naturalist 119: 156-164. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
The aligned sequence data is presented in Nexus format.
(TXT)