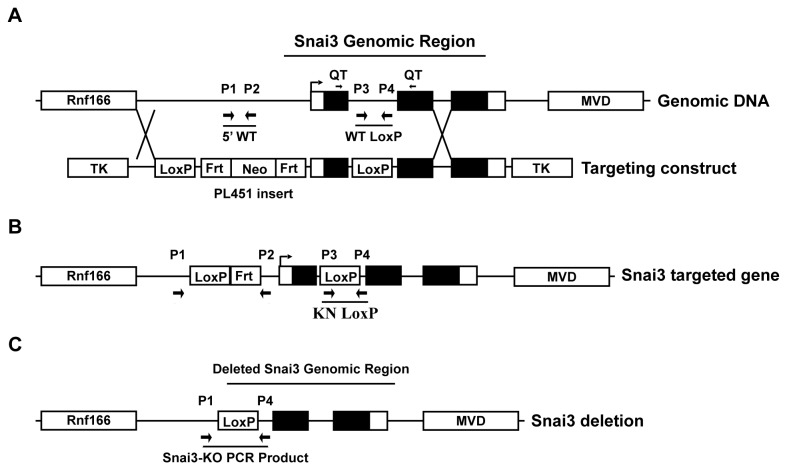
Figure 1. Conditional Snai3 deletion strategy.
(A) The Snai3 WT genomic DNA region contains the Rnf166 gene 5 kb upstream and the MVD gene 10 kb downstream. The Snai3 gene consists of 5’ and 3’ untranslated regions (white boxes), three exons (black boxes), and two introns. Arrow marks the transcriptional start site (TSS). Primers used for genotyping mice and for QT-PCR of Snai3 transcript are labeled as P1-P4 and QT, respectfully, and listed in Supplemental Table 1. The Targeting Construct had a LoxP site inserted into the PacI site of intron one and the PL451 cassette inserted into the SalI site 2kb upstream of the TSS. (B) Homologous recombination created the Snai3 targeted genome containing unique 5’ PL451 and Knockin LoxP PCR products. The Neomycin (Neo) cassette was deleted via FLP-mediated recombination of Frt sites. (C) Cre recombinase activity deleted the Snai3 genomic region and created a new PCR product by bringing together primers P1 and P4, which are normally 6kb apart and unable to make a PCR product. Figure is not to scale.