Cell proliferation and differentiation are two intimately connected fundamental processes. For instance, proliferating neural precursors that undergo terminal neuronal differentiation exit the cell cycle in an irreversible manner, a phenomenon we call mitotic exit. Although we have a good understanding of the gene expression changes that occur during cell cycle arrest, much less is known about the exact molecular mechanisms that enable a permanent shut down of cell cycle gene transcription during mitotic exit. The study by Andrusiak et al.1 sheds some light on this important question.
Transcription factors of the E2F family play a central role in controlling proliferation: in cells that are growing, E2F proteins turn on the expression of genes enabling cell division, while they contribute to shutting down the transcription of those genes in cells that are exiting the cell cycle. The transcriptional regulatory function of E2F proteins comes in part from their ability to recruit protein complexes that help alter the structure of chromatin. In dividing cells, E2Fs can recruit histone methyltransferases or acetyltransferases to impart chromatin marks that facilitate gene transcription, such as H3K4me3 or H3ac (reviewed in ref. 2). On the other hand, in cells that have stopped to divide, E2Fs interact with one of the pocket proteins, the retinoblastoma protein pRb or its relatives, p107 and p130. By binding to E2Fs, the pocket proteins can passively repress gene expression by blocking E2F transcriptional activation functions. Moreover, they can also actively repress E2F target gene expression by recruiting transcriptional co-repressors that participate in modifying the structure of chromatin (reviewed in ref. 3).
Many of these chromatin remodeling proteins, such as the histone deacetylase HDAC1 or the heterochromatin protein HP1, contain a short stretch of amino acids matching the consensus LXCXE (where X represents any residue), which allows them to interact with the pocket domains of pocket proteins at a site different from the E2F-binding domain. In cultured cells, pRb mutants that lack the LXCXE-interaction domain fail to engage in active transcriptional repression of E2F targets4 and to properly establish pericentric heterochromatin.5 Nevertheless, these mutants retain their ability to block E2F-dependent transcriptional activation and to induce cell cycle arrest.
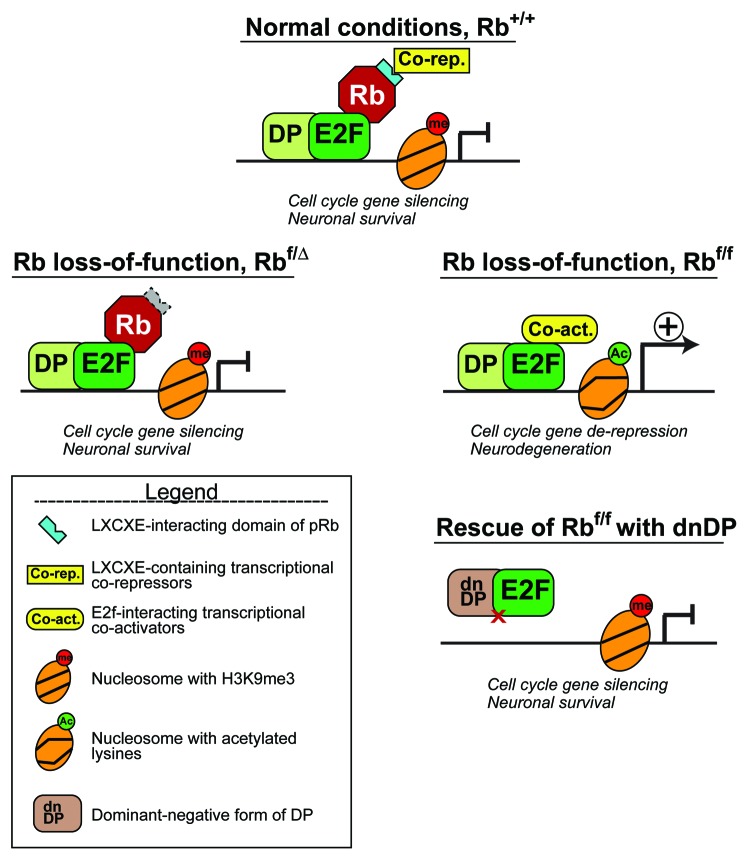
In the brain, Rb is essential to the cell cycle exit and survival of post-mitotic cortical neurons.6 Andrusiak et al. have now examined whether the role of pRb in enforcing the post-mitotic state depends on its capacity to recruit LXCXE domain-containing chromatin remodeling activities.1 They made use of inducible loss-of-function alleles of Rb in vivo and in vitro to prevent the emergence of compensatory effects that can occur with constitutive loss-of-function models. The authors show that while the acute deletion of Rb leads to de-repression of E2F-targeted cell cycle genes in cortical neurons, those genes remain silent when the only remaining allele of Rb codes for a protein unable to interact with LXCXE domain-containing proteins (Fig. 1). Moreover, they show that the complete elimination of Rb changes the chromatin from a state refractory to transcription (nucleosomes trimethylated on H3K9 and poorly acetylated) to one that is more conducive to gene expression. Finally, the authors demonstrate that E2F proteins are responsible for the change in chromatin structure after complete deletion of Rb: no change in chromatin structure takes place when E2F proteins are prevented from binding to their target genes, and cells remain in their post-mitotic state. This rescue experiment demonstrates that pRb keeps mitotically arrested neurons out of the cell cycle by blocking E2F-mediated transcriptional activation of cell cycle genes, rather than by recruiting LXCXE-containing chromatin remodeling complexes.
Figure 1. LXCXE-independent chromatin remodeling and cell cycle repression in mitotically arrested cells. Andrusiak et al. reported that in cortical neurons of the adult brain, which have permanently ceased proliferation, the acute loss of pRb leads to chromatin remodeling at cell cycle genes, to cell cycle re-entry and to apoptotic loss of neurons. In contrast, loss of pRb’s ability to interact with LXCXE-containing proteins does not lead to changes in chromatin structure or to cell cycle re-entry. Moreover, the loss of pRb phenotype can be reversed if the action of E2F proteins is blocked by the overexpression of a dominant-negative form of DP1 that blocks E2F DNA binding.
These findings inform us on the multiple roles played by pRb in regulating gene silencing. During senescence or after exposure to genotoxic stress, pRb may cooperate with LXCXE-containing proteins to block the expression of target genes and suppress tumorigenesis.7,8 However, permanent mitotic exit appears to rely on a different function of pRb: that of antagonizing E2F-mediated cell cycle gene activation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25071
References
- 1.Andrusiak MG, Vandenbosch R, Dick FA, Park DS, Slack RS. LXCXE-independent chromatin remodeling by Rb/E2f mediates neuronal quiescence. Cell Cycle. 2013;12:1416–23. doi: 10.4161/cc.24527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–62. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan HM, Smith L, La Thangue NB. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene. 2001;20:6152–63. doi: 10.1038/sj.onc.1204793. [DOI] [PubMed] [Google Scholar]
- 5.Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–71. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrusiak MG, Vandenbosch R, Park DS, Slack RS. The retinoblastoma protein is essential for survival of postmitotic neurons. J Neurosci. 2012;32:14809–14. doi: 10.1523/JNEUROSCI.1912-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgo RJ, Thangavel C, Ertel A, Bergseid J, McClendon AK, Wilkens L, et al. RB restricts DNA damage-initiated tumorigenesis through an LXCXE-dependent mechanism of transcriptional control. Mol Cell. 2011;43:663–72. doi: 10.1016/j.molcel.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]