Figure 2.
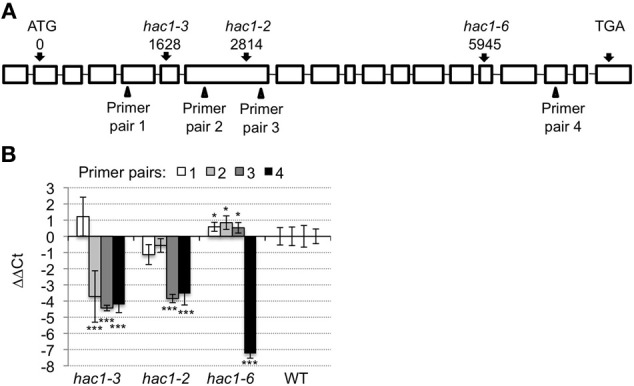
HAC1 T-DNA locations and affects on HAC1 expression. (A) Boxes indicate exons and horizontal lines indicate introns in the HAC1. The locations of the T-DNA inserts in the HAC1 genomic sequence (AT1G79000.2) are indicated with respect to the first nucleotide of the start codon. The positions of the four primer pairs used for the qRT-PCR analyses depicted in panel (B) are indicated by triangles. (B) Mutant and wild-type (WT) Arabidopsis seeds were surface sterilized and then incubated in the dark at 4°C for 3 days prior to being sown on nytex screens on Petri plates containing solid minimal Arabidopsis media. The seeds were incubated under continuous light at room temperature for 20 h, and then the nytex screens and seeds were transferred to Petri plates containing Arabidopsis minimal media supplemented with 0.1 M sorbitol. After an additional 12–13 h, seeds were harvested, followed by isolation of RNA. Quantitative RT-PCR was then used to measure transcript levels. The transcript levels of ACT7 and UBQ6 were determined and the geometric means of their Ct used to normalize transcript levels. Transcript levels are expressed as ΔΔCt. ΔΔCt = ΔCtindicated pimer pair in wild-type – ΔCtindicated primer pair in indicated line. ΔCt = Ctindicated primer pair in indicated line – CtACT7/UBQ6 in same line. Positive ΔΔCt values indicate that HAC1 transcript levels appear higher in the indicated hac1 line than in wild-type when measured using the indicated primer pair. Conversely, negative ΔΔCt values indicate that ACT7 transcript levels appear lower in the indicated hac1 line than in wild-type when measured using the indicated primer pair. Two technical replicates were performed for each biological replicate. Error bars indicate standard deviations. For each primer pair, the Student's t-test was used to compare HAC1 transcript levels in each hac1 mutant line vs. wild-type. *p < 0.1; or ***p < 0.02, according to a Student's t-test. N = 4.
