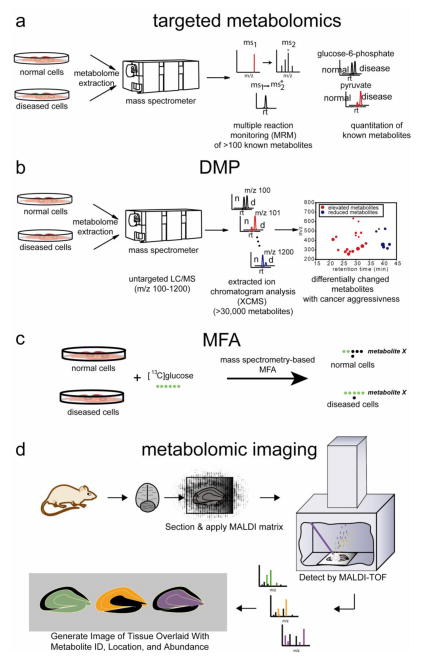
Figure 3. Metabolomic approaches for investigating metabolism.
a) For targeted metabolomics, specific known metabolites are targeted by commonly through multiple reaction monitoring (MRM) by LC-MS/MS, which measures the abundance of a metabolite-specific fragment (ms2) that arises from fragmentation of its corresponding parent mass (ms1). b) For untargeted or DMP-based metabolomics, the mass spectrometer scans a large mass range (m/z 100–1200) in an unbiased manner for both known and unknown metabolites. The resulting large datasets are then analyzed by bioinformatics programs (e.g. XCMS), which aligns, quantifies, and identifies metabolites that are significantly altered between two comparison groups. (n=normal, d=disease) c) For metabolic flux analysis, cells or mice can be treated with isotopic tracers such as [13C]glucose or [13C]glutamine and MRM-based LC-MS/MS or GC/MS can be used to quantify 13C-incorporation rates into cellular metabolites.