Fig. 1.
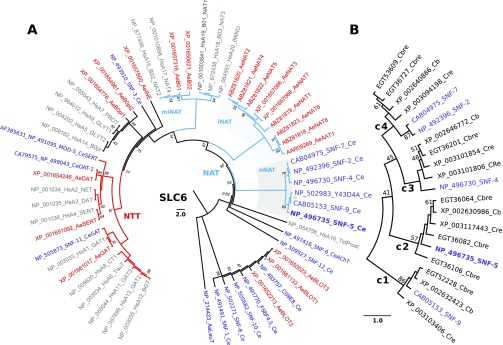
Relationships of nematode nutrient amino acid transporters (NATs) in a phylogenomic tree of the SLC6 family. (A) The evolutionary history was inferred using the UPGMA method (Sneath and Sokal, 1973) based on complete sets of SLC6 members from human (Hs), mosquito Aedes aegypti (Ae) and nematode Caenorhabditis elegans (Ce) genomes (also indicated by different colors). The NCBI protein accession numbers, followed by the name of the transporters from HUGO or WormBase (Version WS234) nomenclature, are shown. For human transporter names, the redundant SLC6 string was removed. The bootstrap consensus tree inferred from 2000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches, except for support values >95%, which were deleted (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter=1). The analysis involved 57 amino acid sequences. The tree was rooted with a sequence of bacterial transporter LeuT (Yamashita et al., 2005). All ambiguous positions were removed for each sequence pair. There were a total of 455 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). (B) The phylogenetic branch of NATs from selected Caenorhabditis genomes: Cb, C. briggsae; Cbre, C. brenneri; Cre, C. remanei. Four paralogous NAT clusters are marked with large numbers 1–4. NAT, nutrient amino acid transporters resembling (n) nematode, (i) insect, and (mi) mammal and insect clusters, also highlighted by gray background; NTT, neurotransmitter transporter.
