Abstract
Cell-free hemoglobin (Hb) resulting from hemolysis is redox-active. It disrupts the oxidoreducing microenvironment, shifting the balance unfavorably towards tissue cytotoxicity. Thus, immediate suppression of the Hb-redox activity and removal of the cell-free plasma Hb is vital for maintaining homeostasis and survival. Among several known Hb-binding plasma proteins, haptoglobin (Hp) is the primary antioxidant of Hb. However, Hb-mediated oxidative stress persists in the covalently bound Hb-Hp aggregates or when Hp is depleted during a severe hemolysis. We therefore inquired whether there is an alternative anti-oxidative defense tactic in the blood. Here, we identified an Hb-interactome in the plasma, constituting Hb, Hp and lipid-free apolipoprotein A-I (apoAI). We found that apoAI acts rapidly as a secondary antioxidant to interact with Hb and quench the Hb-redox activity. We showed that apoAI either acts independently or collaborates with Hp to downregulate Hb-redox activity. Following the association between apoAI and Hb, the apoAI facilitates the uptake of Hb by interacting with a scavenger receptor class B type 1 (SR-BI) displayed on macrophages and hepatocytes. Our findings suggest that apoAI mediates an efficient pathway for the elimination of cytotoxic Hb redox activity.
Keywords: Antioxidant; apolipoprotein A-I; redox-active hemoglobin; scavenger receptor class B type 1 (SR-BI)
Introduction
Hemoglobin (Hb) is a major iron-containing metalloprotein normally sequestered in the erythrocytes. However, cell-free Hb that is liberated as a result of infection by hemolytic microbes, or trauma/ internal hemorrhage, or medication is highly cytotoxic due to its redox-activity. Oxidation of the heme-iron moiety in the Hb initiates a pseudoperoxidase (POX) catalytic cycle, resulting in the production of highly reactive free radicals [1]. The trigger of Hb-POX activity can either elicit a localized antimicrobial activity during an infection [2,3], or induce oxidative stress and damage in many circumstances [4]. Studies have shown that the release of massive levels of Hb might lead to hemolysis-associated smooth muscle dystonia, vasculopathy, and endothelial dysfunction [5]. Moreover, the interrelationship between reactive oxygen species (ROS) and innate immune system in acute lung injury [6], chronic granulomatous disease [7], hemorrhagic shock and ischemia [8] has been reported. Thus, the activated Hb becomes systemically offensive to the host as its avalanche of free radicals imbalances our redox status and impairs our immune responsive blood cells.
It is therefore crucial to rapidly suppress such redox reactivity and remove the cytotoxic Hb from circulation, for which plasma proteins are envisaged to play an important role. Several plasma proteins like haptoglobin (Hp), albumin and transferrin have been reported to bind Hb, scavenging either the free radicals generated or the dissociated free heme [9,10]. Amongst these proteins, Hp is known to be the primary Hb-binding protein, functioning as an antioxidant to attenuate the redox-active Hb [11]. However, during an excessive hemolysis or transfusion of Hb [12], or if Hb-Hp covalently aggregates [13], the Hp is exhausted, leading to the persistence of Hb-driven oxidative stress. Hence, we inquired whether the human body would have additional means to safeguard against Hb-cytotoxicity in such challenging circumstances. This prompted us to isolate potential alternative Hb-interacting partners (Hb-interactome) from the plasma and to track their fate.
Our initial pull-down study using Hbα1 and Hbβ subunits as baits isolated Hp, lipid-free apolipoprotein A-I, and fibrinogen, indicating the presence of a potential Hb-interactome in the human plasma. Apolipoprotein A-I is the major component of the high-density lipoprotein (HDL) [14]. However, the lipid-free form of apolipoprotein A-I (henceforth referred as apoAI) has been implicated to play pathophysiological roles [15]. The association of Hp with the apoAI (on HDL) was found to influence the HDL-mediated transport of cholesterol [16]. Hp was reported to protect the structure and function of apoAI against hydroxyl radicals during acute phase inflammation [17,18]. Furthermore, Hp-related proteins and apolipoproteins have been reported to possess anti-parasitic activity when together with Hb [19], implying a functional importance realized through the liaison of these proteins. These collective observations prompted us to investigate the functional significance of the interactions among Hb, Hp and apoAI.
We found that upon association with Hb, the lipid-free apoAI acts as an antioxidant to decrease the redox-activity of Hb. It can function either independently or in collaboration with Hp. The Hb-apoAI complex was internalized by macrophages and hepatocytes through apoAI-mediated recognition of a cell surface scavenger receptor class B type 1 (SR-BI), suggesting a novel receptor-mediated clearance of Hb from the plasma. Endocytosis of the Hb-apoAI complex induced pro- and anti-inflammatory cytokines. Thus, in addition to the well-known Hb-Hp-CD163 receptor-mediated pathway [20], our findings support the action of apoAI in an alternative route to remove the cytotoxic Hb from the plasma. The apoAI-mediated pathway is likely to be crucial when Hp is depleted in trauma / severe hemolysis, or when Hb-Hp aggregates.
Materials and methods
Biologicals and biochemicals
All chemicals used were of high quality molecular biological grade. Purified human hemoglobin (metHb), haptoglobin 1-1 (Hp), superoxide dismutase, catalase, albumin and transferrin (both from human plasma) were from Sigma-Aldrich. cDNA recombinant clones of human scavenger receptor class B member 1 transcript variant 1 (SCARBI or SR-BI) was obtained from Origene (USA). Human plasma was obtained from the Blood Donation Center/Transfusion Service, National University Hospital, Singapore, under National and Institutional guidelines on ethics and biosafety (Institutional Review Board, Reference Code: NUS-IRB 06-149). Biacore CM5 sensor chips, glutathione Sepharose and rProtein A Sepharose were from GE Healthcare. Primary antibodies raised in rabbits: anti-Hb, anti-Hp, anti-fibrinogen, anti-actin, were purchased from Sigma. Mouse anti-apoAI, mouse anti-Hb, rabbit anti-SR-BI, goat anti-ATP-binding cassette transporter 1 (ABCA1) and rabbit anti-transferrin receptor were from Santa Cruz Biotechnology. Secondary antibodies: goat anti-rabbit HRP conjugate and goat anti-mouse HRP conjugate were from DAKO (Denmark). ZyMax Streptavidin HRP conjugate, anti-mouse Alexa Fluor 488 conjugate, Streptavidin Alexa Fluor 647 conjugate and Alexa Fluor 568-conjugated goat anti-rabbit IgG were from Invitrogen. Prolong Gold antifade reagent with DAPI was purchased from Invitrogen. Sulfo-EGS crosslinker, Sulfo-NHS-SS-Biotin and EZ-Link Sulfo-NHS-SS-Biotinylation Kit were from Thermo Scientific (USA). ABTS chromogen substrate solution for ELISA was from Invitrogen. Proximity Ligation Assay kit was obtained from OLINK Bioscience (Sweden).
E. coli expression vectors, pGEX-4T-1 (Amersham, ampicillinR) and pET16b (Novagen, ampicillinR), were used for constructing the glutathione S-transferase (GST) fused and His-tag fused recombinant proteins, respectively. E. coli Top10 and BL-21 (DE3) strains were used for cloning and expression of recombinant constructs of pGEX-4T-1 and pET16b, respectively. Cell lines, HepG2 and THP-1, were obtained from ATCC, USA. DMEM and RPMI culture media, fetal bovine serum and penicillin-streptomycin solutions were from Invitrogen, Gibco.
Cloning, expression and purification of GST fused recombinant Hbα1 and Hbβ
The E. coli expression vector, pGEX-4T-1, was used for constructing the recombinant clones. The human hemoglobin cDNA clones (Accession Numbers: BC101846 and BC007075, encoding Hbα1 and Hbβ, respectively) purchased from Open Biosystems (USA) were used as templates for PCR amplification. The forward primer: 5’ CGGGATCCG (BamHI) -GTGCTGTCTCCTGCCGACA AG 3’ and the reverse primer: 5’ CGGAATTC (EcoRI) -TTAACGGTATTTGGAGGTCAG 3’ were used to amplify the 426-bp Hbα1 while the forward primer: 5’>ACGCGTCGACTC (SalI) -GTGCA CCTGACTCCTGAGGA <3’ and the reverse primer: 5’>ATAAGAATGCGGCCGC (NotI) -TTAGTGATACT TGTGGGCCA<3’ were used to amplify the 441-bp Hbβ. The recombinant expression constructs were verified by DNA sequencing, and transformed into E. coli BL-21 (DE3) for the protein expression. Induced expression of recombinant proteins were performed according to the manufacturer’s instruction (GST Gene Fusion System). Overnight cultures of the respective clones were separately inoculated into 2x YT broth supplemented with 100 μg/ml ampicillin and 0.3 mM heme precursor, δ-aminolevulinate (Sigma). The clones were grown at 30°C with shaking until the OD600 reached 0.5 - 0.6. The cultures were then induced with 0.2 mM IPTG for an additional 5 h. Both of the recombinant Hbα1 and Hbβ proteins were soluble in the cell lysate. The recombinant GST-Hbα1 and GST-Hbβ fusion proteins were purified by using glutathione Sepharose affinity chromatography (GE Healthcare) according to the manufacturer’s instructions. The purified proteins were electrophoretically resolved on 10% Tris-Tricine SDS-PAGE, and verified by immuno-blotting. Purified GST alone was used as a negative control for the experiments.
GST pull-down
GST-fused recombinant Hbα1 and Hbβ were constructed, expressed and purified separately. Purified GST, GST-Hbα1 and GST-Hbβ were then conjugated to glutathione Sepharose in phosphate buffered saline (PBS) before the assay. Simultaneously, human plasma was pre-cleared using glutathione Sepharose at 4°C for 2 h. 50 μg of each recombinant subunit-conjugated Sepharose was then mixed with 1 ml of pre-cleared plasma in each reaction to pull down plasma proteins at 4°C overnight. The Sepharose with bound proteins were collected by a brief spin and washed thrice with PBS. The bound proteins were eluted with 30 μl of 10 mM glutathione in PBS for 10 min at room temperature (RT) and analysed on 10% Tris-Tricine SDS-PAGE. Protein bands were identified by MALDI-TOF-TOF mass spectrometry. To activate the recombinant Hb subunits [2], the GST-Hbα1 or GST-Hbβ protein bound on Sepharose was partially cleaved by V8 protease from Staphylococcus aureus (Thermo Scientific, >500 Units/mg protein) at a protease:protein ratio of 1:200 (0.25 Unit:50 μg) for 30 min at RT. The Sepharose-conjugated partially cleaved GST-Hb proteins were also used for the pull-down experiment.
MALDI-TOF-TOF Mass Spectrometry
The protein bands of interest, separated on SDS-PAGE, were excised and in-gel digested with trypsin (Promega) according to Shevchenko et al [21]. The trypsinized peptide samples were analyzed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) with Voyager-DE STR Biospectrometry Workstation (Applied Systems) in the Proteins and Proteomics Centre, National University of Singapore. The peptide mass and sequence were analyzed using Matrix Sciences Mascot search at www.matrixscience.com/search_form_select.html.
Co-immunoprecipitation (Co-IP)
Both human plasma and cell extracts were pre-cleared by incubation with rProtein A Sepharose at 4°C for 2 h. The metHb solution was freshly prepared in PBS. For Co-IP with human plasma, two different amounts (500 or 750 μg) of the metHb and protease inhibitor cocktail (Roche) were reconstituted with 500 μl of the pre-cleared plasma to facilitate protein interactions at 4°C for 2 h. For Co-IP with cell extracts, protease inhibitor cocktail was freshly supplemented prior to the addition of primary antibodies. One μg each of the respective primary antibodies was added into each reaction and incubation was resumed at 4°C for an additional 2 h. The resulting protein-antibody complexes were then pulled down using 30 μl of rProtein A Sepharose slurry at 4°C for 1 h with gentle agitation. The protein complex bound on Sepharose was washed thrice before elution with 20 μl of 0.1 M glycine-HCl, pH 2.5, into tubes containing 10 μl of 1 M Tris-HCl, pH 8. The protein solutions were analyzed on SDS-PAGE followed by immunoblotting with their corresponding antibodies.
Cloning, expression and purification of His-tagged recombinant human apoAI
The E. coli expression vector, pET16b, was used for constructing the recombinant human apolipoprotein A-I (apoAI). Human apoAI cDNA clones (Accession Numbers: BC110286) purchased from Open Biosystems (USA) was used as a template for PCR amplification. The forward primer: 5’>GGAATTCCATATG (NdeI)-AAAGCTGCG GTCTGACCTTG<3’ and the reverse primer: 5’>CGCGGATCC (BamHI)-CTGGGTGTTGAGCTTCT TAGT<3’ were used to amplify the 801-bp apoAI. The recombinant expression constructs were verified by DNA sequencing, and transformed into E. coli BL-21 (DE3) for the recombinant protein expression. The expression and purification of recombinant apoAI proteins were performed according to Ryan et al [22]. Briefly, single colonies were picked from overnight bacterial plate culture and inoculated into NCZYM media supplemented with 100 μg/ml ampicillin followed by incubation at 37°C until OD600 reached 0.5 - 0.6. The culture was then induced with 0.1 mM IPTG for an additional 3 h. The harvested bacterial cells were used for the purification of recombinant apoAI (apoAI). To dissociate any lipid that might be associated to the apoAI protein, further purification was carried out under denaturing condition of 3 M guanidine hydrochloride followed by TALON® polyhistidine-tagged affinity chromatography (Clontech) according to the manufacturer’s instructions. The purified denatured apoAI was refolded and buffer-exchanged by using the Amicon Ultracentrifugal filter device (Millipore). The refolded proteins were electrophoretically resolved on 10% Tris-Tricine SDS-PAGE, and verified by immuno-blotting.
End-point solid-phase binding ELISA
His-tagged recombinant human apoAI (apoAI) was purified as a lipid-free form of apoAI. One μg of metHb or Hp 1-1 or apoAI in PBS was separately coated onto maxisorp 96-well plates at 4°C overnight. Unoccupied sites were blocked with 5% BSA in PBST (0.05% Tween 20 supplemented PBS) at 37°C for 2 h. Then, 1- 4 μg of the respective proteins of interest were added and incubated at 37°C for 2 h. The bound proteins were detected by using their respective primary antibodies (anti-Hb or Hp or apoAI) and secondary antibodies (goat anti-rabbit HRP conjugate or goat anti-mouse HRP conjugate) at 37°C for 2 h. Three washes with PBST were introduced after each step of incubation, followed by the addition of ABTS chromogen substrate to detect the HRP conjugate, and the colorimetric reading was recorded at OD415 nm. The mean readings from triplicate reactions for each of the three independent binding experiments were plotted.
Surface Plasmon Resonance (SPR) analysis using Biacore 2000
To characterize the binding kinetics, each protein of interest was immobilized on CM5 chips according to the manufacturer's protocol. Flow cell #1 in each chip, without immobilization of any ligand, was used as a reference surface. PBS supplemented with 5 mM dithiothreitol and 0.005% Tween-20 was used as running buffer at a flow rate of 30 μl/ min. A series of concentrations ranging from 0.1 to 1.6 μM for each protein was injected onto the chip to monitor specific protein-protein interaction for a period of 180 sec, followed by 300 sec dissociation time. Injection of buffer alone served as a negative control. 10 μl of 0.1 M NaOH was used as a regeneration buffer. Data obtained were double-referenced against the reference surface as well as the negative control (buffer alone), and analyzed using BIAevaluation 4.1. The data were fitted with a 1:1 binding model, and the binding affinity was calculated. Only values with Chi2 <10 were deemed acceptable.
Chemiluminescence assay for Hb-POX cycle activity
A superoxide substrate, Cypridina luciferin analog (CLA) [23,24], was used in the chemiluminescence assay (CLA-CL) to track the production of superoxide, which represents the instant activation of the Hb-POX cycle [2]. A reaction mixture of 100 μl in PBS, pH 7.3 was assembled with 20 μM CLA, 5 mM H2O2 and different combinations of metHb, Hp, apoAI, superoxide dismutase (SOD), and catalase (CAT). SOD served as a control for the specificity of CLA used as the substrate for superoxide, while CAT was used as a control for demonstrating the activation of metHb-POX due to its oxidation by H2O2. Proteins were mixed before luminescence recording to ensure the instantaneous complex formation. The reading was immediately taken at 1-sec intervals for a period of 150 sec by using GloMaxTM 20/20 luminometer (Promega). The superoxide produced was measured in relative luminescence units per second (RLU.S-1). The rate of superoxide production was calculated from the linear portion of the CLA-CL curve. The percentage changes were obtained by normalizing the data against the negative control containing metHb alone.
Binding of Hb, apoAI, Hp or their complexes on the cell surface
2 x 106 HepG2 cells were freshly seeded onto each 60 mm culture dish overnight. 4 x 106 THP-1 cells were activated into macrophages by treatment with 30 ng/ml phorbol myristic acid (PMA, Sigma) for 24 h before the experiment. The following steps were carried out at RT. The cells were washed thrice with binding solution containing DMEM or RPMI, supplemented with 0.2% BSA. 1 μM each of metHb, Hp, apoAI or their complexes (pre-complexed for 30 min at RT) was added to allow binding of the respective proteins/ complexes on the cell surface for 30 min and washed to remove any unbound proteins. Cells incubated with binding solution alone served as untreated negative control. The membrane receptor-bound proteins were cross-linked using 2 mM ethylene glycolbis (sulfosuccinimidylsuccinate, sulfo-EGS; Pierce), according to the manufacturer’s instructions. Crosslinking was necessary for subsequent analysis of the composition of the proteins that might be tethered to the cell membrane [25,26]. The cells were harvested and the membrane protein fraction was extracted using ProteoJET membrane extraction kit (Fermentas). The membrane protein fractions were analyzed on SDS-PAGE followed by immunoblotting. For Co-IP of the membrane extract of cells bound with the proteins of interest, the same procedures were carried out according to the protocols described earlier where the anti-apoAI or anti-SR-BI was used.
Proximity Ligation Assay (PLA)
To directly visualize protein-receptor interaction on cell surfaces in situ, the DuolinkTM PLA kit was used. 1 x 104 HepG2 cells or activated THP-1 macrophages cultured in 8-well chamber slides (Iwaki) were washed and bound with apoAI or metHb-apoAI (pre-complexed for 30 min at RT) according to the procedures described earlier. After removal of unbound proteins, the cells were fixed with 4% (w/v) paraformaldehyde-PBS for 10 min followed by blocking with 10% fetal bovine serum (FBS) in PBS for 2 h. The bound proteins were probed with paired primary antibodies of rabbit anti-SR-BI (1:500 diluted) and mouse anti-apoAI (1:1000 diluted), or rabbit anti-SR-BI and mouse anti-Hp (1:2000 diluted). These primary antibodies were then recognized by paired secondary antibodies conjugated with oligonucleotides - anti-rabbit PLA probe MINUS and anti-mouse PLA probe PLUS. In principle, the two oligonucleotides present in the two PLA probes will hybridize if they are in close proximity (as in protein-protein interaction) during the hybridization step. Following that, the two hybridized oligonucleotides were ligated by Duolink ligase, amplified by Duolink Polymerase and detected by red fluorescent-labeled oligonucleotides, which are visible under fluorescent microscope. The procedures for detection were according to the manufacturer’s instructions. PLA signals were visualized using LSM 510 Meta Confocal Laser Scanning system (Carl Zeiss). All images were taken through a 100x oil immersion lens.
To determine the specificity of the interaction between SR-BI and apoAI/Hb, the coated cells were washed thrice and pre-treated with 0.5 μg of anti-SR-BI. After incubation at 37°C for 1 h, the cells were washed thrice to remove unbound antibody prior to binding with the respective proteins. Subsequent steps were as described above.
Quantification of protein internalization by biotin labeling
The procedures for biotin labeling and quantification of cell surface proteins were carried out according to Turvy and Blum [27]. Sulfosuccinimidyl-2-(biotinamido) ethyl-1,3-dithiopropionate (sulfo-NHS-S-S-biotin; Pierce) was used for biotinylation. Briefly, metHb and apoAI were biotin-labeled according to the manufacturer’s instructions. The same amounts of biotinylated metHb and apoAI were incubated at RT for 30 min to allow complexation. The cell surface receptors of 8 x 106 HepG2 cells or activated THP-1 macrophages, seeded on 100 mm culture dish, were also biotin-labeled. The labeled cells were incubated with the labeled proteins or labeled complexes at 37°C for up to 30 min. Biotin-labeled cells incubated with buffer alone served as negative controls. Subsequently, the cells were snap-chilled on ice to halt the protein internalization, and the remaining biotin label on the cell surface was stripped. The cells were lysed on ice for 30 min. A control of untreated cells labeled with biotin but without stripping, was included for quantification of the total cell surface biotin-labeled receptors. The protein concentrations of cell lysates were determined by Bradford assay. Following this, sandwich ELISA was used to quantify the endocytosed biotinylated receptors or proteins. 1 μg each of antibodies against transferrin receptor (TFR), ATP-binding cassette transporter type 1 (ABCA1), SR-BI, Hb or apoAI, diluted in 100 μl PBS was immobilized separately on 96-well maxisorp immunoassay plates at 4°C overnight. The surfaces were blocked with 5% FBS in PBS for 1 h and the same amount of cell lysate containing endocytosed biotin-labeled receptors or proteins was added into each well. Each sample was tested in triplicates. The biotin-labeled protein bound on the plate was detected by HRP-Streptavidin Conjugate (ZyMaxTM Grade, Invitrogen). ABTS substrate enabled the detection of the HRP conjugate and OD415 nm was read. Three washes with PBST were carried out between incubations. The level of endocytosed receptors was calculated as a percentage of the total biotin-labeled cell surface proteins, while the readings for the total amount of biotin-labeled metHb, apoAI or metHb-apoAI used for incubation, were used to calculate the percentage of endocytosed proteins. To prove the specificity of SR-BI receptor-mediated endocytosis on the Hb-apoAI, we blocked the formation of clathrin-coated pits by treating HepG2 cells with 0.45 M sucrose prior to biotinylation, and the cells were maintained in sucrose at 37°C for subsequent incubation with the proteins for endocytosis study.
Immunofluorescence microscopy
To track the localization of the proteins, immunofluorescence microscopy was performed. MetHb dissolved in Hank's Buffered Salt Solution (HBSS) was biotin-labeled by using EZ-link Sulfo-NHS-SS-Biotinylation kit, according to the manufacturer’s instructions. The same amounts of biotin-labeled metHb and apoAI were incubated at RT for 30 min to allow complexation, followed by cross-linking with sulfo-EGS. 1 x 105 HepG2 cells or the activated THP-1 macrophages were freshly seeded onto 13 mm cover slips placed in a 12-well plate and cultured overnight. The cells attached on cover slips were briefly washed twice with pre-warmed HBSS and incubated with the biotin-labeled metHb-apoAI complex at 37°C for 30 min. Unbound protein complexes were then removed by washing with PBS followed by fixation with 4% paraformaldehyde-PBS for 10 min. The cells were permeabilized with 0.05% Triton X-100 in PBS for 5 min and blocked with 10% FBS-PBS overnight at 4°C. The internalized proteins were probed with the respective primary antibodies, anti-apoAI (diluted 1:500) and anti-SR-BI (diluted 1:100), followed by the secondary fluorescence antibodies: Alexa Fluor 488-conjugated goat anti-mouse IgG (diluted 1:1200), Alexa Fluor 568-conjugated goat anti-rabbit IgG (diluted 1:400 for THP-1 and 1:800 for HepG2) and Streptavidin Alexa Fluor 647 conjugate (diluted 1:2500 for THP-1 and 1:3000 for HepG2). The antibodies were diluted in PBS containing 1% FBS. After each incubation step, 3 washes with PBST were performed. The cover slips were dehydrated in 95% and 100% ethanol for 2 and 3 min, respectively, before mounting with ProLong® Gold antifade reagent with DAPI. The sample was visualized on LSM 510 Meta Confocal Laser Scanning system (Carl Zeiss). All images were taken with a 100x oil immersion lens. A control involving sucrose treatment was performed to block the receptor-mediated endocytosis (described earlier).
Determination of cytokine production
2.5 x 105 HepG2 cells or the activated THP-1 macrophages were cultured in 24-well plates overnight before the experiment. The cells were washed once and incubated with nonsupplemented DMEM or RPMI, respectively. Three different amounts: 0.01, 0.05 and 0.1 mg/ml of metHb alone, apoAI alone or metHb-apoAI (pre-complexed), were incubated for 30 min at RT and then added into each well followed by incubation at 37°C for 1 h. The culture was refreshed with the respective media supplemented with 1% FBS for an additional 24 h. The culture supernatant was collected after centrifugation at 500 g for 5 min and stored at - 30°C before assay. Each sample was analyzed in triplicates. BD OptEIA™ Human IL-6 and TNF-α ELISA Kits were used to determine the cytokines produced by the treated cells. Three independent tests were performed for each experiment. Every assay was done in triplicates. For qualitative analyses, a typical representative result was presented; for the quantitative data, the means ± s.d. from data of 3 independent tests was presented. Statistical analyses were performed using two-tailed student’s t-test.
Results
Hb pulls down plasma Hp, apoAI, and fibrinogen
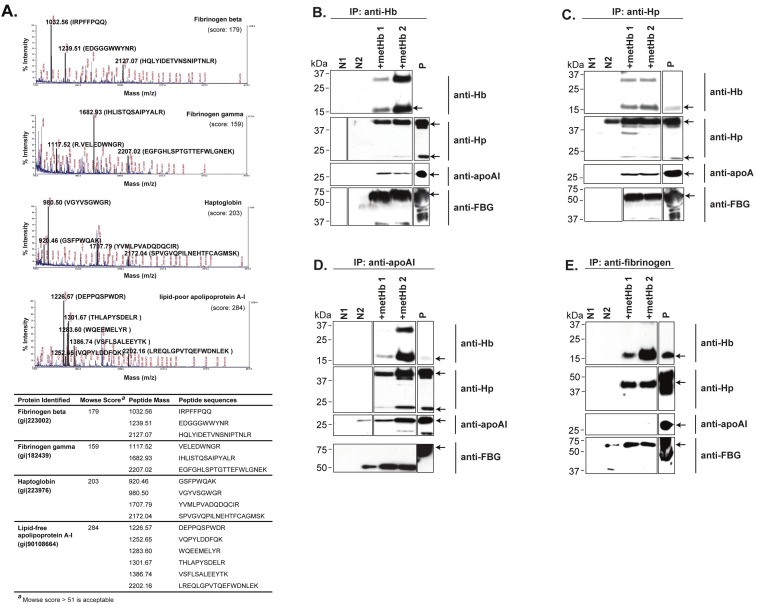
Recombinant Hb subunits (GST-fused Hbα1 and Hbβ), purified through glutathione Sepharose affinity chromatography, were bound to Sepharose and either untreated or treated with V8 protease, resulting in native or proteolytically-activated Hb, respectively. These two possible forms of Hb are deemed to exist in the plasma during hemolytic bacterial infection [2]. The result showed that both the untreated and V8-treated Hb subunits pulled down similar proteins (data not shown), suggesting that the plasma proteins interact indiscriminately with different forms of cell-free Hb. Mass spectrometric analysis identified the pulled down proteins to be Hp, lipid-free apoAI, and β and γ chains of fibrinogen (FBG) (Figure 1A). Hp is known to bind the β-subunit of Hb [28]. Interestingly, our results showed that apoAI only binds to the Hbα1 subunit.
Figure 1.
Identification of plasma proteins which interact with Hb. (A) Mass spectra of Hb-interacting proteins obtained from GST pull-down. The Hb-interacting plasma proteins were isolated by GST pull-down, resolved on SDS-PAGE and identified by MALDI-TOF-TOF mass spectrometry. The matched peptides, their corresponding peaks and protein ID are highlighted and listed. The respective Mowse score for each protein identified are indicated, all of which are significantly greater than the cutoff value of 51. (B-E) Co-IP of Hb-interactome. Two independent experiments performed with different sets of metHb (lanes metHb 1 and 2) showed reproducible results. Proteins recognized by their respective antibodies (arrows): Hb, 16 kDa; Hp, 45 & 18 kDa; apoAI, 28 kDa; fibrinogen (FBG), 60 kDa. Negative control N1: human plasma incubated with rProtein A Sepharose; N2: human plasma incubated with respective antibodies and rProtein A Sepharose (since human plasma contains Hp, apoAI and FBG, these proteins would be pulled down by their respective antibodies during Co-IP and recognized by Western blot on lane N2). Positive control P: metHb probed with anti-Hb, or human plasma probed with other antibodies. The boxes demarcate the images either taken from different lanes of the same blot or from a different blot.
By Co-IP, we found that apoAI, FBG, Hb and Hp were present in the pull-down products when either anti-Hb or anti-Hp was used as bait (Figure 1B, C). However, FBG was not pulled down by anti-apoAI, and apoAI was not pulled down by anti-FBG (Figure 1D, E), indicating no direct interaction between apoAI and FBG, but implying two independent complexes amongst the four proteins (Hb-Hp-apoAI & Hb-Hp-FBG). Since the liaison of Hb, Hp and apoAI has implications on the pathophysiological state, we focused our attention on the Hb-Hp-apoAI interactome.
Lipid-free apoAI interacts with Hb and Hp
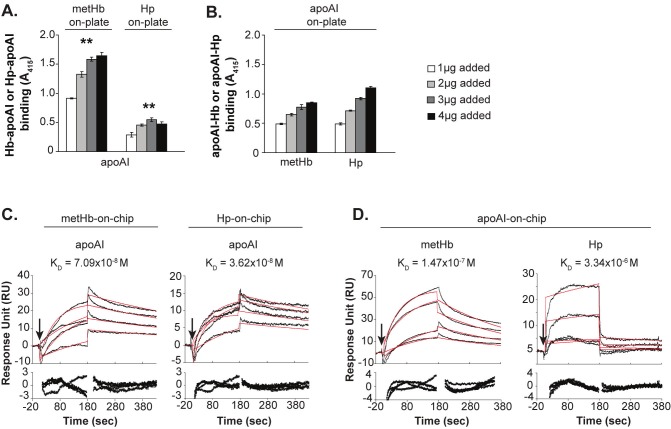
End-point ELISA and real-time SPR analyses were employed to reaffirm the Hb-Hp-apoAI interaction. The assays used His-tagged lipid-free recombinant apoAI (apoAI). ELISA showed that apoAI bind dose-dependently (**) to the immobilized metHb or Hp (Figure 2A). When apoAI was immobilized on the plate, a similar trend of dose-dependent binding of metHb or Hp to the apoAI was observed (Figure 2B). Consistent with the ELISA result, the SPR analysis showed that apoAI bound strongly and dose-dependently to the metHb-on-chip or Hp-on-chip (Figure 2C). Similarly, immobilization of apoAI caused metHb and Hp to bind dose-responsively (Figure 2D) with affinities ranging from 10-6 to 10-8 M (Table 1). Our results from pull down of human plasma proteins (Figure 1A), ELISA and SPR (Figure 2) experiments all corroborate the ability of lipid-free apoAI to interact with cell-free Hb or Hp. Henceforth, we used lipid-free recombinant apoAI to further characterize the physiological consequence of Hb-apoAI interaction.
Figure 2.
Verification of Hb, Hp and apoAI interactions. (A, B) ELISA binding assays for metHb, Hp, and apoAI. Absorbance at 415 nm, representing the amount of protein bound onto the coated plates, was plotted. ** indicates statistical significance (p<0.05) calculated based on student t-test. Data are means ± s.d. of 3 independent experiments. (C, D) Sensorgrams in response unit (RU) of the binding among metHb, Hp, and apoAI. Black curves are the actual binding responses; red curves are the best-fit responses. The arrows indicate the point of injection of sample. The residual plots shown below each sensorgram demonstrate any potential deviation between the fitted curve and the experimental data. The SPR results are reliable since the residual plots are at/near the basal zero level.
Table 1.
Kinetics analysis of binding among Hb, Hp, and apoAI
| Ligand immobilized | Analyte injected | ka (1/Ms) | kd (1/s) | KD (M) | Chi2 |
|---|---|---|---|---|---|
| metHb-on-chip | apoAI | 2.17 x 104 | 1.54 x 10-3 | 7.09 x 10-8 | 0.747 |
| Hp-on-chip | apoAI | 3.64 x 104 | 1.32 x 10-3 | 3.62 x 10-8 | 0.733 |
| apoAI-on-chip | metHb | 2.08 x 104 | 3.61 x 10-3 | 1.47 x 10-7 | 1.56 |
| Hp1-1 | 4.28 x 102 | 1.43 x 10-3 | 3.34 x 10-6 | 5.44 |
ApoAI curbs the redox-reactivity of Hb
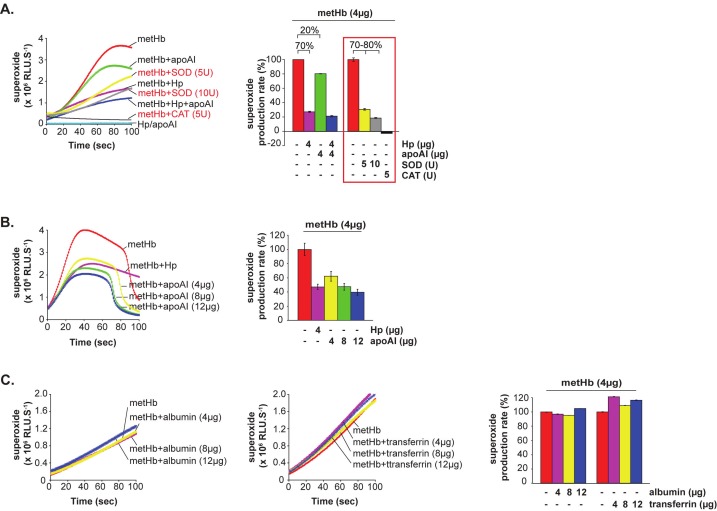
To determine the physiological significance of Hb-apoAI interaction, we inquired whether and how apoAI might affect the redox activity of metHb (HbFeIII), which is the major form of cytotoxic Hb [1]. By using chemiluminescence CLA assay which detects superoxides [2], we tracked the POX activity of metHb upon its association with Hp or apoAI. We found that binding of apoAI to metHb reduced superoxide production by 20%, compared to the positive control, Hp, which suppressed the superoxide generation by ~70% (Figure 3A). Thus, apoAI might play a secondary role as an antioxidant against the Hb. In the presence of SOD, the production of superoxide was inhibited dose-dependently by 70-80% (Figure 3A, red text/box), verifying that CLA is the specific substrate for superoxide in this assay. Furthermore, CAT completely abrogated the production of superoxide, confirming that H2O2 was responsible for triggering the metHb-POX activity.
Figure 3.
Effects of apoAI and Hp on the superoxide production by metHb. CLA was used as a specific substrate for superoxide produced by metHb. The production of superoxide (line chart) is designated as relative luminescence units (RLU.S-1) and the linear portion of the CLA curves was used to calculate and re-plot the production rate of superoxide (%, bar chart). 4 μg of metHb was used for each reaction. (A) CLA-CL assay of metHb in the presence of Hp and/or apoAI, with or without superoxide dismutase (SOD) or catalase (CAT). Red text/box highlights the reactions in the presence of SOD or CAT. (B) CLA-CL assay of metHb in the presence of increasing amounts of apoAI. 4 μg Hp was used as a positive control for suppression of metHb-POX activity. (C) CLA-CL assay of metHb in the presence of increasing amounts of albumin or transferrin (used as negative controls).
Increasing the concentration of apoAI added to metHb progressively decreased the superoxide production rate of metHb. At 2-3 fold concentration (8-12 μg), the apoAI suppressed the superoxide production as effectively as that of Hp (Figure 3B), suggesting that lipid-free apoAI could be a dominant antioxidant when Hp is absent or depleted. Albumin and transferrin, which are plasma proteins known to interact with Hb, were used as controls. Increasing amounts of these two proteins did not inhibit the metHb-mediated production of superoxide (Figure 3C). This corroborates the specificity of the antioxidant action of apoAI against the Hb upon their association.
Physiological significance of the antioxidant activity of apoAI in the plasma
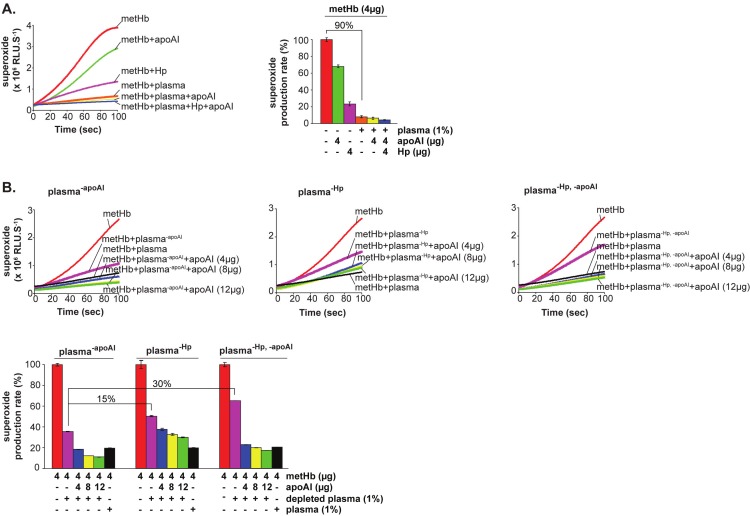
The biological significance of the antioxidant activity of apoAI was further tested in the presence of whole plasma or plasma depleted of Hp or apoAI or both. We found that 1% human plasma significantly decreased the redox activity of metHb by up to 90%, which was much stronger than that of the presence of apoAI or Hp alone (Figure 4A). This is not surprising since it is known that plasma contains large amounts of Hp and other antioxidants [29-31]. However, plasma supplemented with additional amounts of purified apoAI alone (Figure 4A, yellow) or both apoAI and Hp (Figure 4A, blue) further reduced the production rate of superoxide, suggesting the additional quantitative contribution of apoAI towards the oxido-suppressive activity. This was further confirmed by the depletion of apoAI or Hp or both from the plasma, which increased the superoxide production of metHb (Figure 4B). This is observed as follows: relative to plasma-apoAI, there was a 15% rise in superoxide production when plasma was depleted of Hp (plasma-Hp) and 30% rise with depletion of both (plasma-Hp,-apoAI) (Figure 4B, bottom-most panel). Notably, reconstitution with increasing amounts of apoAI to these depleted plasma samples restored their antioxidant activities, as shown by the decreasing trend in superoxide production (Figure 4B - blue, yellow, green). We envisage that in vivo, in the presence of both Hp and apoAI, the plasma would elicit strong antioxidant activity against the Hb-POX. In the case where Hp is depleted, apoAI is able to suppress Hb redox activity effectively.
Figure 4.
The production of superoxide by metHb is suppressed by plasma and is partially restored by depletion of apoAI and Hp. CLA was used as a specific substrate for superoxide produced by metHb. The production of superoxide (line chart) is designated as relative luminescence units (RLU.S-1) and the linear portion of the CLA curves was used to calculate and re-plot the production rate of superoxide (%, bar chart). 1% of human plasma (with or without depletion of the indicated proteins) and 4 μg of metHb were used in the respective reactions. (A) CLA-CL assay of metHb in the presence of human plasma, Hp and/or apoAI. (B) CLA-CL assay of metHb in the presence of depleted human plasma and repletion with increasing amounts of apoAI. Plasma-Hp and plasma-apoAI represent the human plasma depleted of Hp alone and apoAI alone, respectively. Plasma-Hp,-apoAI indicates plasma depleted of both Hp and apoAI. Immuno-depletion was performed with the respective antibodies.
Hb-apoAI complex binds to SR-BI receptor
To elucidate how the potent redox-active Hb-apoAI complex might be eliminated from circulation, we explored the possible binding of the complex to SR-BI since lipid-free apoAI is a known ligand of SR-BI [14], which clears the oxidized or modified LDL [32]. Thus, THP-1 and HepG2 cells, which express SR-BI [33,34] and are at the vicinity of Hb-apoAI complexes in vivo, were employed to characterize potential interaction between Hb-apoAI and SR-BI.
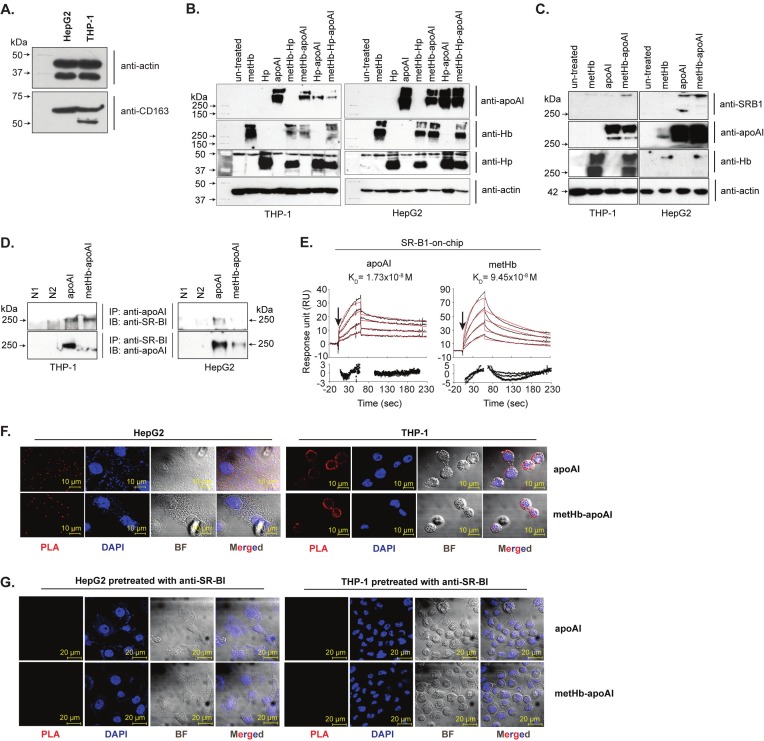
First, we demonstrated that both THP-1 and HepG2 cells do not express a 130 kDa functional CD163 receptor (Figure 5A). Then, immunoblot analyses of the membrane extract showed that only metHb and/or apoAI, but not Hp, was bound to the cell surfaces as complexes larger than 250 kDa (Figure 5B). The absence of CD163 in these cells and the lack of involvement of Hp in the membrane complex fortify our reasoning that an alternative receptor, independent of both CD163 and Hp, plays a role in the binding and elimination of Hb-apoAI complex from circulation. Furthermore, the specificity of such binding was verified using a negative control cell line, HEK293T, which is devoid of endogenous SR-BI. Incubation of Hb-apoAI with HEK293T cells did not result in any protein complexes >250 kDa in the membrane extract (data not shown).
Figure 5.
SR-BI-mediated binding of Hb-apoAI complex. (A) Western blot detection of CD163 receptor expression on HepG2 and THP-1 cells. Both of the cell membrane fractions were extracted and probed with anti-CD163 (Santa Cruz). No functional CD163 of 130 kDa was observed. (B) Immunoblot analysis of membrane extracts of cells incubated with metHb, Hp, apoAI or their complexes and probed separately by anti-apoAI, anti-Hb or anti-Hp to detect the presence of the respective proteins bound on the cell surfaces. (C) Immunoblot analysis of membrane extracts of cells incubated with the metHb, apoAI or their complexes and probed separately with anti-SR-B1, anti-apoAI and anti-Hb. Actin (42 kDa) probed with anti-actin, was used as a loading control. (D) Co-IP of cell membrane extracts incubated with apoAI and metHb-apoAI complex. N1 and N2 are two negative controls. N1 denotes the reaction of membrane extract of untreated cells incubated with rProtein A sepharose; N2 denotes the reaction of membrane extracts of untreated cells incubated with rProtein A Sepharose and anti-apoAI, or with rProtein A sepharose and anti-SR-BI. (E) Sensorgrams and residual plots (see legend to Figure 2) of SPR analyses of binding kinetics between apoAI, metHb and immobilized recombinant SR-BI. Black curves, actual binding responses; red curves, best-fit responses. (F) In situ PLA of cells incubated with apoAI or metHb-apoAI complex. PLA signals appear as red dots, each of them represents a pair of interactive proteins; nucleus, stained by DAPI (blue). The cells incubated with apoAI or metHb-apoAI complex were probed with anti-apoAI, anti-SR-BI and PLA, respectively. (G) Anti-SR-BI pre-treatment prior to the binding of apoAI or metHb-apoAI. No red PLA signals were found on the cells surface, confirming the specific interaction of apoAI and SR-BI.
Western blot showed that the >250 kDa protein complexes contained SR-BI, metHb and apoAI (Figure 5C), indicating their potential interactions. However, Co-IP could only confirm the binding of apoAI and SR-BI (Figure 5D) because of the cross-reactivity between anti-Hb and anti-SR-BI. Nevertheless, SPR analysis showed that both apoAI and metHb bind dose-responsively to the immobilized SR-BI at KD of 1.73-9.45x10-8 M (Figure 5E, Table 2), with apoAI displaying higher affinity for SR-BI, which is consistent with apoAI being the natural ligand of SR-BI.
Table 2.
Kinetics analysis of binding of Hb and apoAI to SR-BI
| Ligand immobilized | Analyte injected | ka (1/Ms) | kd (1/s) | KD (M) | Chi2 |
|---|---|---|---|---|---|
| SR-BI-on-chip | metHb | 5.94 x 104 | 5.61 x 10-3 | 9.45 x 10-8 | 5.01 |
| apoAI | 6.38 x 104 | 1.11 x 10-3 | 1.73 x 10-8 | 0.627 |
The binding of Hb-apoAI complex to the cells via recognition between apoAI and SR-BI was visualized by in situ Proximity Ligation Assay (PLA). Protein-receptor interactions indicated by red fluorescent signals were observed on the cells bound with apoAI or metHb-apoAI (Figure 5F), demonstrating the interaction between apoAI and SR-BI. The specificity of interaction was shown by the lack of PLA signal on cells incubated with Hp or PLA probe alone (data not shown). In addition, cells pretreated with anti-SR-BI antibody abolished binding of the proteins (Figure 5G). These results confirm that SR-BI is the receptor recognizing the Hb-apoAI complex, via specific interaction with apoAI.
SR-BI mediates internalization of Hb-apoAI complex
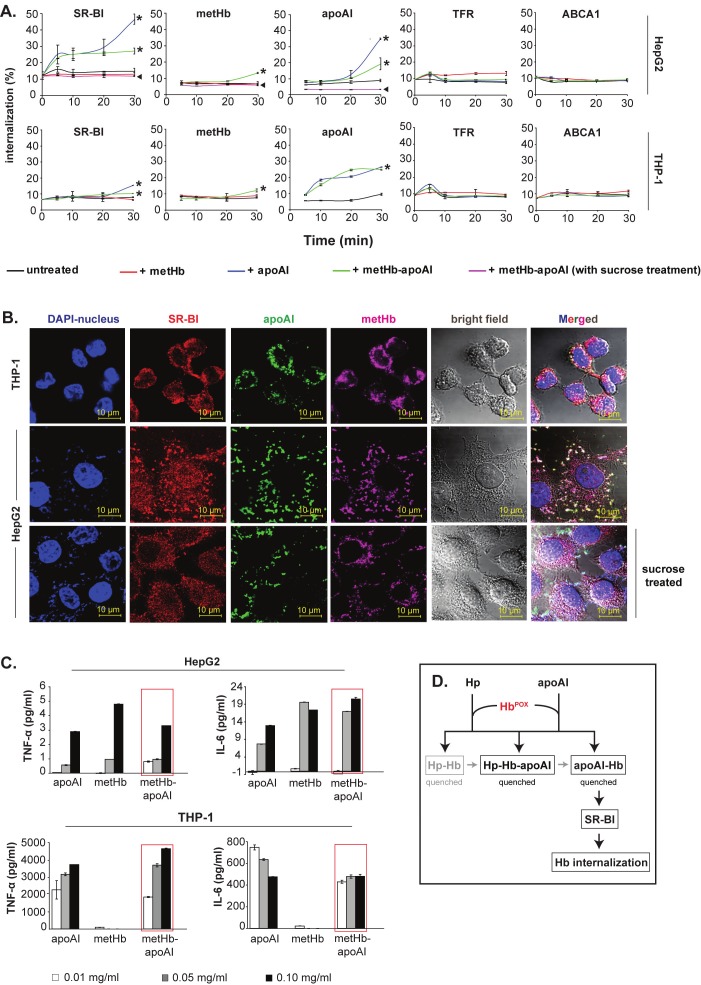
Following confirmation of the anchorage of Hb-apoAI onto SR-BI, we quantified the receptor-mediated internalization of the complex. Over 30 min, we observed dynamic internalization of SR-BI together with metHb-apoAI complex into the HepG2 and THP-1 cells (Figure 6A, *), but not with negative control receptors, TFR and ABCA1 (Figure 6A). The SR-BI receptor-mediated internalization of the Hb-apoAI complex was impaired by sucrose treatment of the cells (Figure 6A, ◄), which prevented the formation of clathrin-coated pits, indicating the specificity of the membrane receptor-mediated uptake of the Hb-apoAI complex. Over the time course studied, no internalization occurred in cells incubated with metHb alone (Figure 6A), suggesting that apoAI is needed to facilitate endocytosis of the Hb-apoAI complex via the SR-BI receptor.
Figure 6.
SR-BI-mediated internalization of Hb-apoAI complex. (A) Quantification of the internalization of SR-BI, metHb and apoAI into hepatocytes (HepG2) and macrophages (THP-1). The % endocytosed proteins were normalized against either total biotinylated cell surface expression for the SR-BI receptor or total biotinylated proteins applied (metHb, apoAI); * indicates the internalized SR-BI, metHb or apoAI; ◄ indicates the internalization level upon treatment with 0.45 M sucrose. Two receptors, TFR and ABCA1, served as negative controls in the cells incubated with metHb and/or apoAI. (B) Immunofluorescence microscopy of internalized metHb-apoAI. The intracellular white spots in the merged images indicate co-localized proteins. (C) Cytokine production by HepG2 and THP-1 following endocytosis of metHb-apoAI. Increasing amounts of proteins (metHb and/or apoAI) were incubated with the cells for 1 h to allow endocytosis to occur. The production of TNF-α and IL-6 in the cell supernatant was determined after 24 h. The red boxes highlight the dose-dependent upregulation of TNF-α and IL-6 upon endocytosis of metHb-apoAI complex. (D) A model to illustrate the antioxidant activities of lipid-free apoAI and Hp. Cell-free oxidative Hb was rapidly captured by Hp and apoAI to form complexes in plasma, all of which quenched the redox activity of Hb (HbPOX). Following association with Hb, the apoAI further leads the internalization of Hb-apoAI complex via its recognition by SR-BI on macrophages and hepatocytes. Black color represents interactions identified in this study while grey indicates either known or speculated process.
Immunofluorescence microscopy aided the visualization of such internalization, where biotinylated metHb-apoAI and SR-BI were observed to co-localize in the cell cytoplasm (Figure 6B, top two panels; white spots in the merged image). Due to its limited area of cytoplasm, THP-1 cells demonstrated a tight white ring around the typically large nucleus of the monocytes. Consistent with the above quantitative result (Figure 6A, ◄), HepG2 cells pre-treated with sucrose also clearly showed loss of uptake (Figure 6B, bottom-most panel; reduced white spots), although some protein binding occurred on the cell membrane.
Dose-dependent upregulation of TNF-α and IL-6 was observed upon endocytosis of metHb-apoAI (Figure 6C, red boxes), indicating immune response upon internalization of Hb-apoAI via SR-BI-positive cells.
Discussion
We identified for the first time, an interaction between lipid-free apoAI and cell-free plasma Hb (in the presence and absence of Hp). Our findings support the existence of an alternative antioxidant, lipid-free apoAI in the plasma, which effectively decreases the Hb-POX activity and further mediates the uptake of the cytotoxic Hb by the macrophages and hepatocytes through the SR-BI receptor. This defense pathway protects tissues from oxidative stress (Figure 6D). While the Hbβ subunit is known to bind Hp [28], our results showed that apoAI only binds the Hbα subunit. This indicates that the Hb tetramer may display different binding sites for the recruitment of the 3 plasma proteins, thus simultaneously forming various complexes: Hb-Hp, Hb-apoAI or Hb-apoAI-Hp. The interaction between lipid-free apoAI with metHb and Hp (ELISA and SPR experiments) corroborates the existence of such complexes (Figure 2).
The association of apoAI with Hb resulted in the decrease of the redox activity of Hb (Figure 3 & 4) indicating that apoAI either independently or in collaboration with Hp, shields us from Hb cytotoxicity in vivo. Furthermore, we showed that on its own, higher amounts of lipid-free apoAI can possibly achieve a level of antioxidant activity similar to that of Hp. Since the lipid-free form of apoAI normally exists transiently and is in low quantity in the plasma [35], we envisage that its antioxidant activity is subtle and strategically timed to target the cell-free pro-oxidative Hb or Hb-Hp aggregates. It is conceivable that the role of lipid-free apoAI in scavenging the prooxidant Hb is particularly crucial when Hp is exhausted during excessive hemolysis, like in hemodialysis patients where increased level of lipid-free apoAI was observed [36]. However, further studies are needed to strengthen this proposal.
We have shown that inconsequential to the well-known route of elimination of plasma Hb via Hp-CD163 pathway, the apoAI facilitates an alternative path of uptake of plasma Hb through specific interaction with SR-BI receptor (Figure 5 & 6). Although apoAI exhibits high affinity for SR-BI (Figure 5E), we cannot preclude possible direct internalization of Hb via SR-BI, albeit at lower rate. Furthermore, internalization of the Hb-apoAI complex could also occur via other apoAI receptors [37,38], besides SR-BI. This remains to be tested in future. Additionally, a comparison study with the known CD163 receptor could provide further insights into SR-BI-mediated internalization of Hb, afterall, both SR-BI and CD163 receptors are present on the macrophages and it is reasonable to envisage that they could act independently or collaboratively to clear the cytotoxic plasma Hb effectively. Nevertheless, the endocytosis of Hb-apoAI complex triggered both pro- and anti-inflammatory immune responses, which recapitulates the pathophysiological significance of the concerted interactions of Hb-apoAI-SR-BI.
Acknowledgements
This work was supported by a Tier 2 grant (T208B3109) from Ministry of Education, Singapore. We thank Cheryl Zixian Tang for help with co-immunoprecipitation and mass spectrometry experiments.
Conflict of interest
Hereby, the authors declare that they do not have any conflict of interest.
References
- 1.Alayash AI, Ryan BA, Eich RF, Olson JS, Cashon RE. Reactions of sperm whale myoglobin with hydrogen peroxide. Effects of distal pocket mutations on the formation and stability of the ferryl intermediate. J Biol Chem. 1999;274:2029–2037. doi: 10.1074/jbc.274.4.2029. [DOI] [PubMed] [Google Scholar]
- 2.Du R, Ho B, Ding JL. Rapid reprogramming of haemoglobin structure-function exposes multiple dual-antimicrobial potencies. EMBO J. 2010;29:632–642. doi: 10.1038/emboj.2009.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang N, Tan NS, Ho B, Ding JL. Respiratory protein-generated reactive oxygen species as an antimicrobial strategy. Nat Immunol. 2007;8:1114–1122. doi: 10.1038/ni1501. [DOI] [PubMed] [Google Scholar]
- 4.Alayash AI, Patel RP, Cashon RE. Redox reactions of hemoglobin and myoglobin: biological and toxicological implications. Antioxid Redox Signal. 2001;3:313–327. doi: 10.1089/152308601300185250. [DOI] [PubMed] [Google Scholar]
- 5.Rother RP, Bell L, Hillmen P, Gladwin MT. The Clinical Sequelae of Intravascular Hemolysis and Extracellular Plasma Hemoglobin. JAMA: The Journal of the American Medical Association. 2005;293:1653–1662. doi: 10.1001/jama.293.13.1653. [DOI] [PubMed] [Google Scholar]
- 6.Xiang M, Fan J, Fan J. Association of Toll-Like Receptor Signaling and Reactive Oxygen Species: A Potential Therapeutic Target for Posttrauma Acute Lung Injury. Mediators of Inflammation. 2010;2010 doi: 10.1155/2010/916425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartl D, Lehmann N, Hoffmann F, Jansson A, Hector A, Notheis G, Roos D, Belohradsky BH, Wintergerst U. Dysregulation of innate immune receptors on neutrophils in chronic granulomatous disease. Journal of Allergy and Clinical Immunology. 2008;121:375–382. e379. doi: 10.1016/j.jaci.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 8.Gill R, Tsung A, Billiar T. Linking oxidative stress to inflammation: Toll-like receptors. Free Radical Biology and Medicine. 2010;48:1121–1132. doi: 10.1016/j.freeradbiomed.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponka P, Beaumont C, Richardson DR. Function and regulation of transferrin and ferritin. Semin Hematol. 1998;35:35–54. [PubMed] [Google Scholar]
- 10.Sadrzadeh SM, Bozorgmehr J. Haptoglobin phenotypes in health and disorders. Am J Clin Pathol. 2004;121(Suppl):S97–104. doi: 10.1309/8GLX5798Y5XHQ0VW. [DOI] [PubMed] [Google Scholar]
- 11.Melamed-Frank M, Lache O, Enav BI, Szafranek T, Levy NS, Ricklis RM, Levy AP. Structure-function analysis of the antioxidant properties of haptoglobin. Blood. 2001;98:3693–3698. doi: 10.1182/blood.v98.13.3693. [DOI] [PubMed] [Google Scholar]
- 12.Lim SK, Kim H, bin Ali A, Lim YK, Wang Y, Chong SM, Costantini F, Baumman H. Increased susceptibility in Hp knockout mice during acute hemolysis. Blood. 1998;92:1870–1877. [PubMed] [Google Scholar]
- 13.Kapralov A, Vlasova II, Feng W, Maeda A, Walson K, Tyurin VA, Huang Z, Aneja RK, Carcillo J, Bayir H, Kagan VE. Peroxidase activity of hemoglobin-haptoglobin complexes: covalent aggregation and oxidative stress in plasma and macrophages. J Biol Chem. 2009;284:30395–30407. doi: 10.1074/jbc.M109.045567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S, Laccotripe M, Huang X, Rigotti A, Zannis VI, Krieger M. Apolipoproteins of HDL can directly mediate binding to the scavenger receptor SR-BI, an HDL receptor that mediates selective lipid uptake. J Lipid Res. 1997;38:1289–1298. [PubMed] [Google Scholar]
- 15.Asztalos BF, Roheim PS. Presence and formation of 'free apolipoprotein A-I-like' particles in human plasma. Arterioscler Thromb Vasc Biol. 1995;15:1419–1423. doi: 10.1161/01.atv.15.9.1419. [DOI] [PubMed] [Google Scholar]
- 16.Balestrieri M, Cigliano L, Simone ML, Dale B, Abrescia P. Haptoglobin inhibits lecithin-cholesterol acyltransferase in human ovarian follicular fluid. Mol Reprod Dev. 2001;59:186–191. doi: 10.1002/mrd.1021. [DOI] [PubMed] [Google Scholar]
- 17.Salvatore A, Cigliano L, Bucci EM, Corpillo D, Velasco S, Carlucci A, Pedone C, Abrescia P. Haptoglobin binding to apolipoprotein A-I prevents damage from hydroxyl radicals on its stimulatory activity of the enzyme lecithin-cholesterol acyl-transferase. Biochemistry. 2007;46:11158–11168. doi: 10.1021/bi7006349. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe J, Grijalva V, Hama S, Barbour K, Berger FG, Navab M, Fogelman AM, Reddy ST. Hemoglobin and its scavenger protein haptoglobin associate with apoA-1-containing particles and influence the inflammatory properties and function of high density lipoprotein. J Biol Chem. 2009;284:18292–18301. doi: 10.1074/jbc.M109.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanhollebeke B, De Muylder G, Nielsen MJ, Pays A, Tebabi P, Dieu M, Raes M, Moestrup SK, Pays E. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- 20.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A, Wilm M, Mann M. Peptide sequencing by mass spectrometry for homology searches and cloning of genes. J Protein Chem. 1997;16:481–490. doi: 10.1023/a:1026361427575. [DOI] [PubMed] [Google Scholar]
- 22.Ryan RO, Forte TM, Oda MN. Optimized bacterial expression of human apolipoprotein A-I. Protein Expr Purif. 2003;27:98–103. doi: 10.1016/s1046-5928(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 23.Kawano T, Pinontoan R, Hosoya H, Muto S. Monoamine-dependent production of reactive oxygen species catalyzed by pseudoperoxidase activity of human hemoglobin. Biosci Biotechnol Biochem. 2002;66:1224–1232. doi: 10.1271/bbb.66.1224. [DOI] [PubMed] [Google Scholar]
- 24.Nakano M. Determination of superoxide radical and singlet oxygen based on chemiluminescence of luciferin analogs. Methods Enzymol. 1990;186:585–591. doi: 10.1016/0076-6879(90)86154-n. [DOI] [PubMed] [Google Scholar]
- 25.Bu G, Williams S, Strickland DK, Schwartz AL. Low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor is an hepatic receptor for tissue-type plasminogen activator. Proc Natl Acad Sci U S A. 1992;89:7427–7431. doi: 10.1073/pnas.89.16.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efrat M, Aviram M. Macrophage paraoxonase 1 (PON1) binding sites. Biochem Biophys Res Commun. 2008;376:105–110. doi: 10.1016/j.bbrc.2008.08.106. [DOI] [PubMed] [Google Scholar]
- 27.Turvy DN, Blum JS. Biotin labeling and quantitation of cell-surface proteins. Curr Protoc Immunol. 2001;Chapter 18 doi: 10.1002/0471142735.im1807s36. Unit 18.7. [DOI] [PubMed] [Google Scholar]
- 28.Yoshioka N, Atassi MZ. Haemoglobin binding with haptoglobin. Localization of the haptoglobin-binding sites on the beta-chain of human haemoglobin by synthetic overlapping peptides encompassing the entire chain. Biochem J. 1986;234:453–456. doi: 10.1042/bj2340453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowman BH, Kurosky A. Haptoglobin: the evolutionary product of duplication, unequal crossing over, and point mutation. Adv Hum Genet. 1982;12:189–261. 453–4. doi: 10.1007/978-1-4615-8315-8_3. [DOI] [PubMed] [Google Scholar]
- 30.Tsuji A, Ikeda N, Nakamura T. Plasma lipids, lipoproteins and apolipoproteins and sudden cardiac death. Int J Legal Med. 1999;112:151–154. doi: 10.1007/s004140050221. [DOI] [PubMed] [Google Scholar]
- 31.Wayner DD, Burton GW, Ingold KU, Barclay LR, Locke SJ. The relative contributions of vitamin E, urate, ascorbate and proteins to the total peroxyl radical-trapping antioxidant activity of human blood plasma. Biochim Biophys Acta. 1987;924:408–419. doi: 10.1016/0304-4165(87)90155-3. [DOI] [PubMed] [Google Scholar]
- 32.Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Gugiu B, Fox PL, Hoff HF, Salomon RG, Hazen SL. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J Biol Chem. 2002;277:38503–38516. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 33.Bocharov AV, Baranova IN, Vishnyakova TG, Remaley AT, Csako G, Thomas F, Patterson AP, Eggerman TL. Targeting of scavenger receptor class B type I by synthetic amphipathic alpha-helical-containing peptides blocks lipopolysaccharide (LPS) uptake and LPS-induced pro-inflammatory cytokine responses in THP-1 monocyte cells. J Biol Chem. 2004;279:36072–36082. doi: 10.1074/jbc.M314264200. [DOI] [PubMed] [Google Scholar]
- 34.Murao K, Yu X, Imachi H, Cao WM, Chen K, Matsumoto K, Nishiuchi T, Wong NC, Ishida T. Hyperglycemia suppresses hepatic scavenger receptor class B type I expression. Am J Physiol Endocrinol Metab. 2008;294:E78–87. doi: 10.1152/ajpendo.00023.2007. [DOI] [PubMed] [Google Scholar]
- 35.Neary RH, Gowland E. Stability of free apolipoprotein A-1 concentration in serum, and its measurement in normal and hyperlipidemic subjects. Clin Chem. 1987;33:1163–1169. [PubMed] [Google Scholar]
- 36.Okubo K, Ikewaki K, Sakai S, Tada N, Kawaguchi Y, Mochizuki S. Abnormal HDL apolipoprotein A-I and A-II kinetics in hemodialysis patients: a stable isotope study. J Am Soc Nephrol. 2004;15:1008–1015. doi: 10.1097/01.asn.0000117286.85443.7d. [DOI] [PubMed] [Google Scholar]
- 37.Hidaka H, Tozuka M, Yamauchi K, Ohta H, Nakayama J, Katsuyama T. Purification and measurement of HDL3-binding proteins in human peripheral blood mononuclear cells. Ann Clin Lab Sci. 2003;33:271–278. [PubMed] [Google Scholar]
- 38.Matsuyama A, Sakai N, Hiraoka H, Hirano K, Yamashita S. Cell surface-expressed moesin-like HDL/apoA-I binding protein promotes cholesterol efflux from human macrophages. J Lipid Res. 2006;47:78–86. doi: 10.1194/jlr.M500425-JLR200. [DOI] [PubMed] [Google Scholar]