Abstract
POU3F4 is a POU domain transcription factor that is required for hearing. In the ear, POU3F4 is essential for mesenchymal remodeling of the bony labyrinth and is the causative gene for DFNX2 human non-syndromic deafness. Ear abnormalities underlie this form of deafness, characterized previously in multiple spontaneous, radiation-induced and transgenic mouse mutants. Here we report three novel mutations in the POU3F4 gene that result in profound hearing loss in both humans and mice. A p.Gln79* mutation was identified in a child from an Israeli family, revealed by massively parallel sequencing (MPS). This strategy demonstrates the strength of MPS for diagnosis with only one affected individual. A second mutation, p.Ile285Argfs*43, was identified by Sanger sequencing. A p.Cys300* mutation was found in an ENU-induced mutant mouse, schwindel (sdl), by positional cloning. The mutation leads to a predicted truncated protein, similar to the human mutations, providing a relevant mouse model. The p.Ile285Argfs*43 and p.Cys300* mutations lead to a shift of Pou3f4 nuclear localization to the cytoplasm, demonstrated in cellular localization studies and in the inner ears of the mutant mice. The discovery of these mutations facilitates a deeper comprehension of the molecular basis of inner ear defects due to mutations in the POU3F4 transcription factor.
Keywords: Hearing loss, cochlea, ENU, DFNX2
Introduction
Deafness is considered to be the most common sensory disorder in humans, with a genetic origin in about half of the cases [Nance, 2003]. Defining the molecular pathomechanisms involved in deafness is essential for defining the processes of how hearing loss begins and progresses, and ultimately, treatment strategies. Identification and characterization of novel mutations in human families and the study of animal models, in particular the mouse, have been invaluable in deciphering such pathomechanisms [Dror and Avraham 2009; Petit and Richardson, 2009]. Both traditional and recently advanced techniques, such as deep sequencing or massively parallel sequencing (MPS), have led to the discovery of hundreds of genes for isolated or non-syndromic hearing loss in both humans and mice with varying inheritance patterns [Brown et al., 2008; Shearer et al., 2011]. Of these, an estimated 1–5% of human hereditary hearing loss is caused by X-linked mutations in different populations [Reardon, 1990; Petersen et al., 2008]. To date, five deafness loci have been mapped to chromosome X, with three of the corresponding genes identified, PRPS1 for DFNX1, [Liu et al., 2010], POU3F4 for DFNX2 [de Kok et al., 1995] and SMPX for DFNX4 [Huebner et al., 2011; Schraders et al., 2011].
DFNX2 (MIM# 304400) is the result of mutations in the POU3F4 (POU domain class 3, transcription factor 4, BRN-4) transcription factor that belongs to subclass III of the POU superfamily. This family of proteins is characterized by a bipartite DNA-binding domain consisting of a POU-specific domain (POUS) and a POU homeodomain (POUHD). Seven POU subclasses are defined by the linker that separates the bipartite binding domain [Phillips and Luisi, 2000]. While each subdomain is structurally autonomous and binding by POUHD is sufficient, affinity is increased by POUS binding. Pou3f4 is involved in several developmental and regulatory pathways. Pou3f4 and Tbx1 interact for mesenchymal signaling in inner ear formation [Braunstein et al., 2008]. Together with EphA4, Pou3f4 regulates spiral ganglion axon fasciculation, essential for appropriate auditory innervation [Coate et al., 2012]. Retinoic acid, Fgf and Hedgehog signaling pathways play a role in the regulation of Pou3f4 enhancer activity, and Pou3f4 enhancers require Pax2 and Sox2 transcription factors [Robert-Moreno et al., 2010]. Pou3f4 is expressed mainly in the developing brain [Alvarez-Bolado et al., 1995], and inner ear [Phippard et al., 1998], as well as pancreatic cells [Hussain et al., 1997]. However, deafness is the only clinical phenotype found in DFNX2 patients, emphasizing the critical role of Pou3f4 in the ear. In humans, many mutations have been found in this gene, including point mutations, small to partial or complete deletions of the gene, and inversions in the coding region [Bitner-Glindzicz et al., 1995; de Kok et al., 1995], as well as deletions and inversions upstream of the coding region [de Kok et al., 1995; de Kok et al., 1996]. The deafness resulting from these mutations affects males, and is clinically heterogeneous, varying from conductive to mixed to sensorineural deafness, with late-onset hearing loss sometimes observed in female carriers [Marlin et al., 2009]. Anatomically, the mutations are associated with partial hypoplasia of the cochlea, enlarged internal acoustic canal and a characteristic stapes gusher upon surgery and stapes fixation [de Kok et al., 1995].
Corresponding loss-of-function mouse mutants have been invaluable in the study of the mechanisms leading to hearing loss and ear abnormalities due to mutations in Pou3f4. The first DFNX2 mouse model was created by gene-targeted mutagenesis, displayed hearing loss and vestibular defects, and implicated Pou3f4 in development of mesenchymal tissues of the inner ear [Phippard et al., 1999]. Another Pou3f4 knock-out exhibiting deafness with normal vestibular function, revealed alterations in cochlear spiral ligament fibrocytes and implicated potassium ion homeostasis in the Pou3f4 pathway [Minowa et al., 1999]. Further support for Pou3f4 function in the inner ear was supplied by spontaneous and radiation-induced mouse mutants, through the similarities and differences in these different alleles [Phippard et al., 2000; Song et al., 2011].
Additional alleles have been instrumental for helping define more phenotypes. Many of these have been created using N-ethyl-N-nitrosourea (ENU), a chemical used to create genetically-altered mice harboring point mutations [Acevedo-Arozena et al., 2008]. Here we report a new Pou3f4 allele, schwindel (sdl), created within the framework of an ENU mutagenesis program [Hrabe de Angelis et al., 2000]. The mutation in its POU homeodomain is predicted to lead to a truncated protein. In parallel, we report two novel human mutations in the POU3F4 gene, predicted to lead to truncated proteins, with mutations in this gene reported for the first time in the Israeli Jewish population. Given the similar nature of these mutations, the Pou3f4sdl mouse serves as a relevant model to further define the molecular mechanisms associated with POU3F4 hereditary hearing loss in humans.
Materials and Methods
Clinical Evaluation
The study was approved by the Helsinki Committee of Tel Aviv University and the National Helsinki Committee for Human Genetic Research of the Israel Ministry of Health. A medical history was collected, including degree of hearing loss, age at onset, progression of hearing impairment, symmetry of hearing impairment, use of hearing aids and cochlear implants, presence of tinnitus, medication, noise exposure, pathologic changes in the ear, other relevant clinical manifestations, and family history. High-resolution temporal bone computed tomography (CT) was used to detect inner ear malformations. Blood was drawn after subjects signed committee-approved consent forms, and genomic DNA was extracted. All subjects were tested by standard Sanger sequencing for the known deafness genes in the Jewish population, based on the relevant phenotype or ethnic background [Brownstein and Avraham, 2009].
Massively Parallel Sequencing (MPS)
Capture libraries were created and MPS and bioinformatics performed as previously described [Brownstein et al., 2011; Horn et al., 2013]. Briefly, exons and the flanking 40 bp into introns of 284 human deafness-associated genes were selected for capture and sequenced on an Illumina HiSeq 2000 Analyzer (HT-Seq Unit, Technion). In order to identify variants, reads were initially aligned to the Genome Reference Consortium Human Build 37 (GRCh37/hg19) using Burrows-Wheeler Aligner (BWA) [Li and Durbin, 2010]. Single nucleotide variants (SNV) and insertion-deletions (indels) were then called from the aligned data using SAMtools (http://samtools.sourceforge.net) and Dindel (Albers et al., 2011], respectively. Copy number variants (CNV) analysis was done using BreakDancer [Chen et al., 2009]. Rare variants were identified by filtering against dbSNP135, the 1000 Genomes Project, the Exome Variant Server (http://evs.gs.washington.edu/EVS/) and classified by predicted effect on the protein using Polyphen2 [Adzhubei et al., 2010], SIFT [Kumar et al., 2009], and Consurf [Ashkenazy et al., 2010]. The nomenclature of the identified novel variants was checked using Mutalyzer (http://www.LOVD.nl/mutalyzer/) and variants were uploaded to the LOVD locus-specific database (www.lovd.nl/POU3F4).
Mice
The founder mouse of the mouse line, provisionally named KAL001, was generated in a large-scale ENU mutagenesis program [Hrabe de Angelis et al., 2000]. Progeny continued for analysis were given the International Committee on Standardized Genetic Nomenclature for Mice approved name and symbol schwindel (sdl). All procedures involving animals were approved, in Germany, by the Committee on Animal Health and Care of the local governmental body of the state of Bavaria and performed in strict compliance with the EEC recommendations for the care and use of laboratory animals (European Communities Council Directive (86/609/EEC) and in Israel, met the guidelines described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee of Tel Aviv University (M-10-087). The colony was maintained on the original C3HeB/FeJ genetic background.
Plasma Potassium Measurements
Blood samples were collected under ether anesthesia from the retrobulbar vein plexus into Li-heparin-coated sample tubes. Plasma and cells were separated by centrifugation within one to two hours after collection (Biofuge Fresco, 5000xg, 10 min at room temperature). Potassium values were measured in plasma samples using the ion-selective electrode unit of a AU400 clinical chemistry analyzer (Olympus), using controls and calibrators provided by Olympus, Germany.
Mouse Genetic Mapping and DNA Sequencing
To map the gene responsible for the deafness phenotype, affected mice on a C3HeB/FeJ background were crossed to C57BL/6 wild-type mice to generate F1 offspring, and the F1 were intercrossed to generate F2 mice. These mice were tested for their hearing threshold and deaf mice mapped using 74 microsatellite markers as previously described [Hertzano et al., 2008]. After defining linkage to chromosome X on a 25.5 cM region, three additional markers were used to define linkage to the 18 cM region containing the Pou3f4 gene. Sanger sequencing was performed using the ABI BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) and an AB 3500xl DNA Genetic Analyzer.
Auditory Function Analysis
Initial auditory function was assessed by a click-box test using a 20 kHz Clickbox (MRC Institute of Hearing Research, Nottingham). For further analysis, ABR was performed on mice at ages post natal day (P)15 and P30. Mice were anaesthetized by IP injection of Avertin (0.015ml/gr mouse), and placed on a heating pad in an acoustic chamber (MAC-1, Industrial Acoustic Company, Bronx, NY, USA). For hearing threshold evaluation, three sub-dermal electrodes: ground, reference and active were placed at the mouse head behind the left ear, behind the right ear and on the forehead, respectively. TDT (Alachua, FL, USA) software (Biosig RZ) and hardware (RZ and MF1 speakers) were used, with a speaker placed 10cm in front of the mouse. Speakers were calibrated using an ACO pacific 7017 microphone. The mice were presented with click stimuli (0.1ms) at increasing levels, ranging between 10dB–90dB in 5dB steps. Each point of measurement was recorded and averaged 512 times, analyzed and threshold was determined as the lowest recognizable ABR response.
Paint-Fill
Paint-filling of inner ears were performed following a standard protocol [Kiernan, 2006]. Briefly, following decapitation of P0 mice, the heads were bissected, the brain was removed and the heads fixed overnight in Bodian’s fixative (75% EtOH, 5% formalin, 5% glacial acetic acid). After fixation, the samples where dehydrated through a gradient of EtOH solutions 75%, 95%, 100% (2 washes of 2 hours were performed in each solution). The samples were cleared overnight in methyl salicylate (Sigma-Aldrich, St. Louis, MO) under a chemical hood. Injection of paint (1% gloss paint in methyl salicylate) into the endolymphatic labyrinth of the inner ear was performed using a pulled glass capillary pipette anchored on a micromanipulator. The paint-filled inner ears were isolated from the skull and photographed under a binocular.
Histology
Whole inner ears were fixed and decalcified using Bouin’s solution (Sigma-Aldrich) for 4h at room temperature. The ears were processed in a Tissue Processor (Leica TP1020), positioned in paraffin blocks with a Histoembedder (Leica, Wetzlar, Germany) and sectioned using a microtome (Leica Jung RM2055). Paraffin serial sections (10 μm) were then dewaxed in xylene and rehydrated. The standard hematoxylin and eosin (H&E) stain procedure was performed.
Scanning Electron Microscopy (SEM)
Inner ears were dissected out in 1x PBS buffer and fixed in 2.5% glutaraldehyde diluted in 1x PBS overnight at 4°C. The organ of Corti was exposed and all membranes removed. Preparation of the samples was performed using the osmium tetroxide-thiocarbohydrazide (OTOTO) method [Hunter-Duvar, 1978], critical point dried and coated with gold at the Faculty of Life Sciences Electron Microscopy Unit and viewed with a JSM 840A scanning electron microscope (Jeol, Tokyo, Japan).
Immunohistochemistry
Inner ear sections were prepared as for histology (without H&E staining) and labeled with antibodies as previously described [Dror et al., 2010]. Myosin VI (green) labels the inner and outer hair cell cytoplasm. NF200 labels the neurofilaments (turquoise) and 4′,6-diamidino-2-phenylindole (DAPI) stains the nuclei (blue). The following antibodies were used: chicken-anti-Pou3f4 antibody, against amino acids 158–179 of the mouse Pou3f4 protein sequence [Coate et al., 2012], 1:10,000; rabbit anti-myosin VI (Proteus BioSciences, Ramona, CA, USA), 1:200; mouse anti-NF200 (Sigma-Aldrich), 1:300.
Cell Transfection and Immunofluorescence
COS-7 cells were grown in DMEM supplemented with 10% FCS and split into 6-well plates one day prior to transfection. The Pou3f4 construct, pCMV6-AC-GFP-Pou3F4 (Origene, Rockville, MD, USA) was mutated using the QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) in order to introduce the human c.583-584delAT and the mouse c.900T>A mutations. Their sequences were verified by Sanger sequencing. The three plasmids were transfected into the cells using Lipofectamine2000 (Invitrogen). The cells were fixed 24 hours after transfection using 4% PFA and labeled using chicken anti-Pou3f4 [Coate et al., 2012], followed by a goat anti-chicken (Molecular Probes, Life Technologies, Carlsbad, CA, USA) and phalloidin (Invitrogen, Life Technologies) incubation. For nuclear staining, cells were incubated with 5 μm DRAQ5™ dye (Abcam, Cambridge, UK) for 10 min at room temperature. Cells were mounted using Fluorescent Mounting Media (Biotec Applications, Rehovot, Israel) and viewed using an SP5 confocal microscope (Leica).
Intensity Correlation Analysis
Intensity correlation analysis was carried out using ImageJ (http://rsb.info.nih.gov/ij/) with an appropriate plug-in included in the package of the Wright Cell Imaging Facility [Li et al., 2004] (http://www.uhnres.utoronto.ca/facilities/wcif/). For each image, the background signal was eliminated by drawing a region of interest within an area occupied by cells. Both images were automatically thresholded with the algorithm incorporated into the ImageJ software. The Mander’s coefficients were calculated for at least 14 cells from 3 independent experiments. All data are expressed as the mean ± SEM. Statistical significance was determined using the one-tailed student’s t-test.
Results
Targeted Capture and Massively Parallel Sequencing Identifies a Nonsense Mutation in the Human POU3F4 Gene
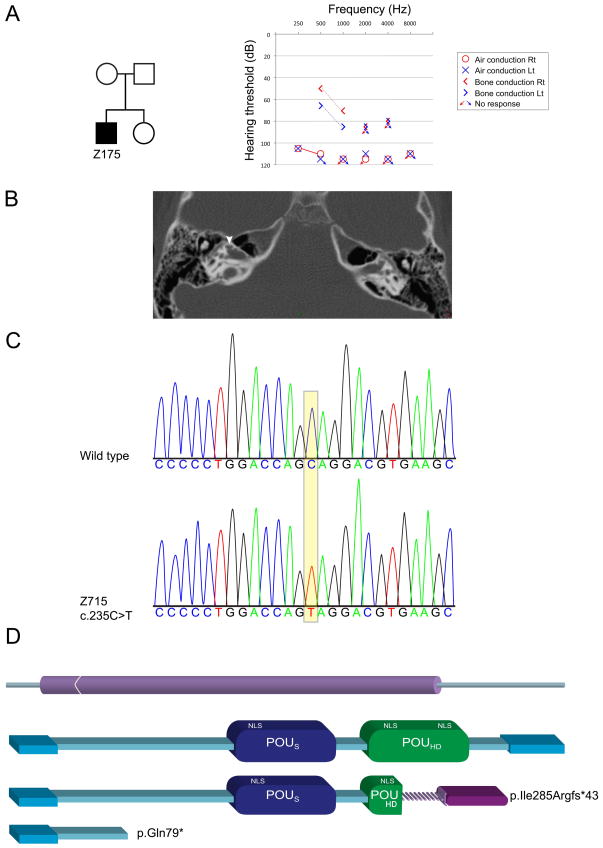
A targeted capture pool we had previously developed for identifying mutations in all known deafness genes was used [Brownstein et al., 2011]. The current pool contained a total of 281 human protein-coding genes, of which 163 were the human orthologues of genes associated with the inner ear or deafness in the mouse, and three human microRNAs [Horn et al., 2013]. One of the probands examined, Z715, a male of Israeli Jewish Ashkenazi descent, has a congenital profound bilateral hearing loss with a Mondini malformation and cochlear implant (Fig. 1A, B). There is no family history by family report. No relevant candidates were detected in the indel and CNV analysis. The only homozygous or compound heterozygote variant candidate recovered in the SNV analysis was in the POU3F4 gene on chrX:82763567, C>T in 25/26=96% reads. The others were nonsynonymous heterozygote variants and were thus disqualified from further analysis. Sanger sequencing confirmed the presence of the variant (Fig. 1B), which corresponds to c.235C>T (NM_000307.3) in the only coding exon, exon 1, and predicted to lead to a stop codon at amino acid 79 out of 361 amino acids, p.Gln79* (NP_000298) (Fig. 1C). The POU3F4 p.Gln79* novel mutation was not detected in the parents and sister of the proband, indicating a de novo mutation. It was also not identified among 211 deaf probands or 145 and 115 hearing controls of Ashkenazi and other Israeli Jewish ancestries, respectively. Following the detection of the first POU3F4 mutation in the Jewish Israeli population, a novel POU3F4 deletion of two base pairs, c.853_854del, was identified in a Jewish family originating from Bulgaria, leading to a predicted frameshift mutation (p.Ile285Argfs*43).
Figure 1.
A POU3F4 nonsense mutation leads to deafness in a male with Mondini dysplasia. A: Pedigree of the family, with one affected child (filled square). The audiogram demonstrates profound sensorineural hearing loss at all frequencies tested. B: An axial CT of proband Z715 shows an incomplete partition type II (Mondini) of both cochleae. The arrow points towards the cystic apex of the cochlea. C: Sanger sequencing chromatogram of a portion of the wild-type human POU3F4 sequence and the corresponding region of the mutation in proband Z715. D: The human mutations are predicted to lead to truncated proteins. The top scheme represents the one-exon structure of the POU3F4 gene. The bottom schemes represent the full-length protein and the predicted truncated forms. The c.853_854del mutation leads to a frameshift and a replacement of the normal C-terminal end of the protein with a new segment, p.Ile285Argfs*43, and the c.235C>T mutation leads to a stop codon p.Gln79*.
Hearing Impairment and Plasma Potassium Levels in ENU Mice
A male G3 animal derived from the recessive mutant screen of the ENU mouse mutagenesis project was shown to have impaired hearing ability, assessed by a click-box test, and a high plasma potassium concentration of 5.5 and 5.8 mmol/l in two subsequent tests within a three-week interval. Further breeding of this animal to a wild-type female and subsequent intercrosses of the offspring resulted in the production of 114 (65 male, 49 female) offspring, out of which only 13 male and 10 female animals showed high potassium values (≥ 5.6 mmol/l), in most cases associated with hearing impairment and head-tossing and occasional circling behavior. A breeding pair, renamed schwindel (sdl), was used to breed offspring for further analyses on the auditory and vestibular phenotype, including the phenotypic characterization and identification of the causative mutation.
An X-linked Nonsense Mutation in the Pou3f4 Gene is Responsible for sdl/sdl
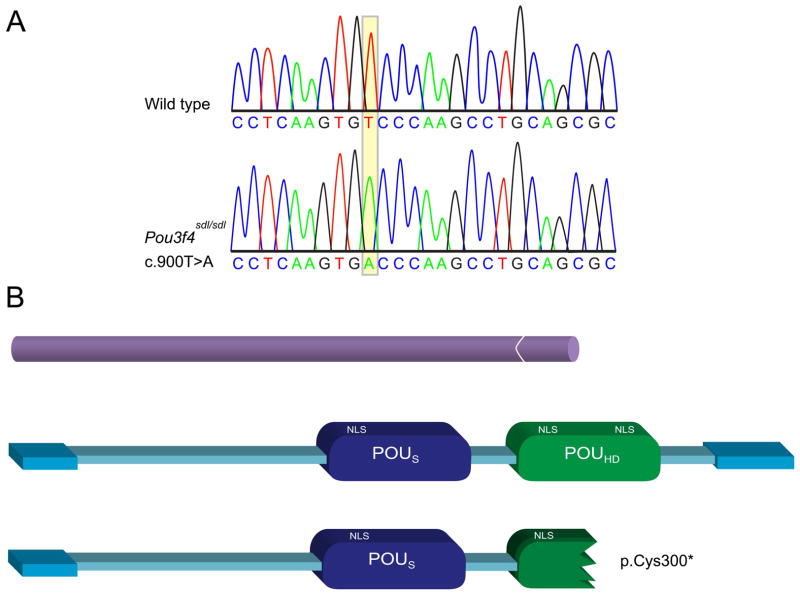
Analysis of the colony mating data provided strong evidence that the sdl phenotype is linked to the X chromosome. These findings were confirmed in a genome scan of 21 F2 deaf mice, revealing linkage of the sdl allele to an 18cM region between markers DXMit16 and DXMit130 on the X chromosome. In a candidate gene approach we favored the Pou3f4 gene, since it is known to be the molecular cause for DFNX2 X-linked recessive deafness in humans and mice [de Kok et al., 1995; Phippard et al., 1999]. Upon sequencing the single exon of the Pou3f4 gene, all 21 backcrossed deaf mutants were found to carry a T to A transversion at nucleotide 900 (c.900T>A, NM_008901.1) (Fig. 2A). The mutation within the POU homeodomain was predicted to cause a premature stop codon at amino acid 300 out of 361 of the Pou3f4 peptide (p.Cys300*, NP_032927) (Fig. 2B). We then sequenced all C3HeB/FeJ-sdl mutants from our original colony and found that the T to A mutation co-segregates with all mice displaying the sdl deafness phenotype. None of the littermate control mice tested carried the T to A change.
Figure 2.
A Pou3f4 nonsense mutation leads to deafness in an ENU-derived mutant mouse, schwindel (sdl). A: Sanger sequencing chromatogram of a portion of the wild-type mouse Pou3f4 sequence and the corresponding region of the mutation in the Pou3f4sdl/Pou3f4sdl mouse. B: The c.900T>A mutation is predicted to lead to a stop codon, p.Cys300*. The upper scheme represents the one exon structure of the Pou3f4 gene. The bottom schemes represent the full-length protein and the predicted truncated form.
Pou3f4sdl Mice Present with Profound Hearing Loss and Impaired Vestibular Function
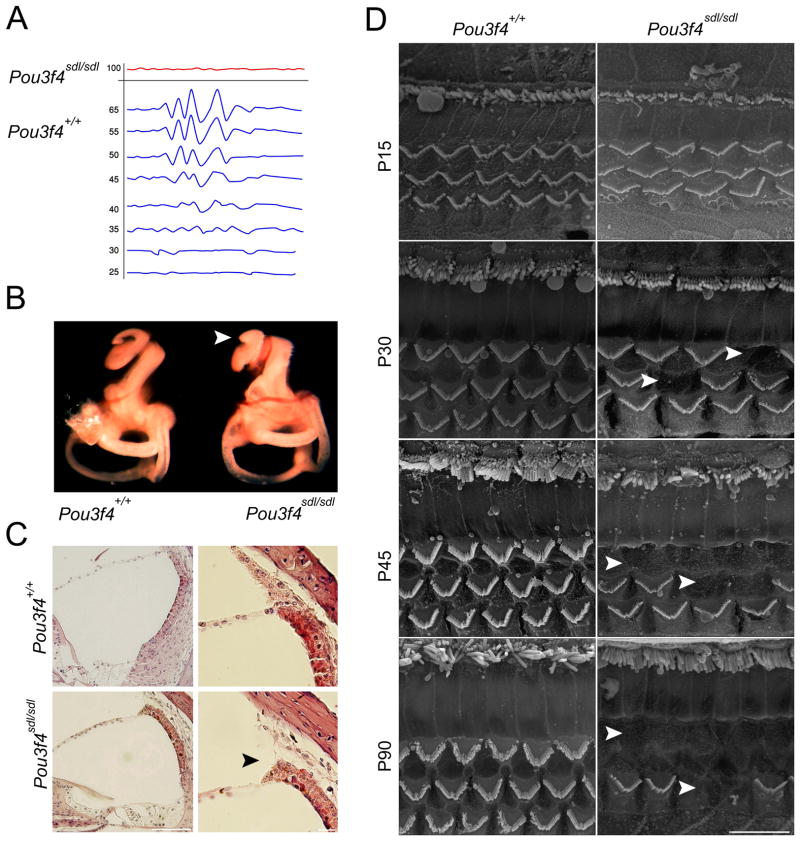
Mutant Pou3f4sdl mice at P30 were initially identified by a click-box test, which elicited a typical startle reflex in wild-type animals, whereas Pou3f4sdl mutant mice did not respond. For further analysis, we performed auditory brainstem response (ABR) measurements on C3H/FeJ-Pou3f4sdl mutant mice and wild-type littermates to quantitatively assess the hearing loss (Fig. 3A). We found that mutant Pou3f4sdl mice had an auditory threshold at a broadband sound stimulus above 90dB, while wild-type mice had a threshold of 30dB (+/−5dB). In addition, the Pou3f4sdl mutants were tested in a battery of vestibular tests including contact righting, tail hang test, observation of circling behavior and swim test and were found to have severe vestibular dysfunction.
Figure 3.
Auditory and morphological characterization of the Pou3f4sdl/Pou3f4sdl inner ears. A: ABR measurements indicate that Pou3f4sdl homozygote mice are deaf at P30. B: Paint-filled inner ears at P0 demonstrate shortening of the cochlear duct by almost one turn. C: Histology and H&E staining demonstrates detachment of the stria vascularis from the spiral ligament. D: SEM at ages P15-P90 demonstrates loss of outer hair cells, evident from P30 onwards.
Pou3f4sdl Inner Ears Present with Subtle Hair Cell Loss that Progresses with Age
Since POU3F4 patients present with structural abnormalities in the cochlear bones [Bitner-Glindzicz et al., 1995], as well as Pou3f4 mutant mice previously studied, for example [Phippard et al., 1998], we evaluated the gross structure of the inner ear using paint fill in cleared ears (Fig. 3B) and H&E staining of paraffin-embedded ear sections (Fig. 3C). The paint-fill analysis revealed shortening of the cochlear duct by almost one turn, similar to a Mondini malformation of the bony labyrinth. H&E staining demonstrated a detachment of the stria vascularis from the spiral ligament. Unlike the stapes abnormalities found in some Pou3f4 mouse mutants, the middle ear bones of Pou3f4sdl mice appeared normal in structure. SEM of the organ of Corti after exposure of the sensory epithelia and removal of the tectorial membrane showed loss of outer hair cells in Pou3f4sdl mice as young as P30 (Fig. 3D). By P45, most of the outer hair cells had degenerated in the base and middle cochlear regions. The innermost row of outer hair cells, closest to the inner hair cells, seemed to be affected first and most severely. We did not observe morphological changes in inner hair cells at any of the ages tested, through P90.
Aberrant Cellular Localization of the Human and Mouse Mutated POU4F3
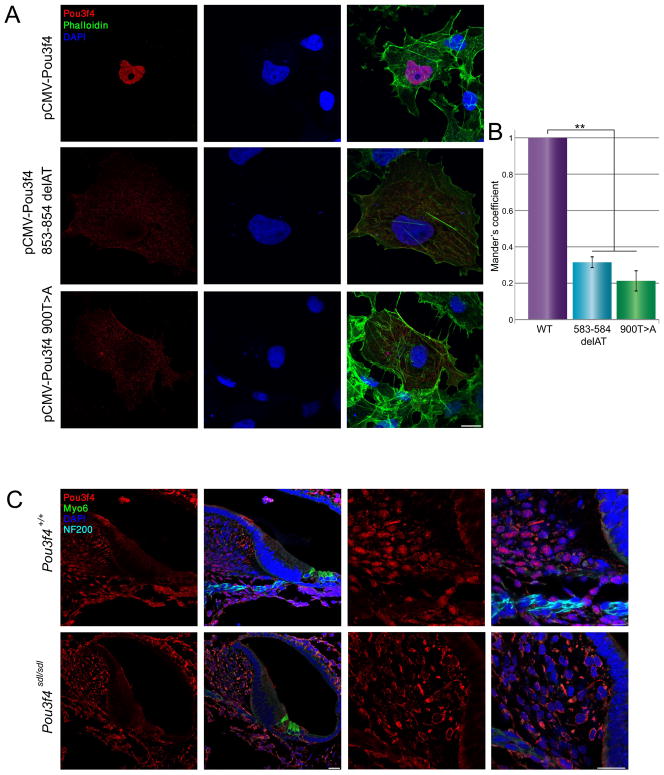
The cellular consequences of the identified mutations were examined by transfecting COS-7 cells with both wild type and mutant forms of human and mouse POU3F4 cDNA (Fig. 4A). Distribution of wild-type Pou3f4 was found in the nuclei of cells, as measured by intensity correlation analysis (0.96 ± 0.01, n = 15), while the human p.Ile285Argfs*43 mutation led to mislocalization of most of the POU3F4 to the cytoplasm in COS-7 cells, with a portion remaining in the nucleus (0.31 ± 0.03, n = 20) (Fig. 4B). Similarily, the p.Cys300* mutation led to an abnormal pattern of expression (Fig. 4B). Pou3f4 expression was diffused mainly to the cytoplasm, with some remaining localization of Pou3f4 in the nucleus (0.21 ± 0.06, n = 14). The human p.Gln79* mutation could not be examined in the same fashion, since the antibody would not recognize the mutant form, given that it is raised against a peptide at amino acids 158–179 in the mouse, corresponding to the same amino acids in humans. In the mouse inner ear, Pou3f4 was detected in the nuclei of cells in the spiral ligament and spiral limbus (Fig. 4C). Inspection of the cellular localization of Pou3f4 in inner ears derived from Pou3f4sdl mice revealed expression of the mutant protein, demonstrating that the mutation does form a protein recognized by the antibody. However, the subcellular localization is altered and the protein is almost exclusively detected in the cytoplasm.
Figure 4.
Both Pou3f4 human and mouse mutations lead to mislocalization of the Pou3f4 protein. A: COS-7 cells transfected with vectors containing Pou3f4 cDNA show wild-type localization in the nucleus, while the mutant forms present mostly in the cytoplasm. B: Intensity correlation analysis demonstrated that in cells transfected with the wild-type Pou3f4 cDNA, Pou3f4 co-localizes completely with DRAQ5, a far-red fluorescent live-cell-permeant DNA dye, whereas in the mutant forms, there is very little co-localization (* p < 0.001). C: Immunofluorescence of Pou3f4 in wild type and Pou3f4sdl inner ears. Pou3f4 (red) is localized in the nuclei, stained by DAPI (blue), of the spiral limbus and fibrocytes. In the mutant, Pou3f4 is absent from the nucleus. Myosin VI (green) labels the inner and outer hair cells. NF200 labels the neurofilaments (turquoise).
Discussion
This study describes novel human POU3F4 mutations resulting in congenital deafness and a mouse Pou3f4 mutation leading to early-onset hearing loss. Characterizing new mouse models that resemble human phenotypes is vital to achieve a complete understanding of specific forms of deafness. Although to date over 60 causative genes for human hereditary non-syndromic hearing loss have been identified (Van Camp and Smith, 2013), genetic diagnosis of affected individuals remains a major challenge. Due to high sequencing costs and low throughput efficiency of traditional sequencing methods, only the most prevalent genes within a target population have been investigated when looking for causative mutations. We have previously shown that the combination of targeted genomic capture and MPS is an efficient method to identify novel and known mutations responsible for hereditary deafness [Brownstein et al., 2011]. We applied this method to determine the causative gene and mutation in a previously undiagnosed deaf individual of an Israeli Jewish family. The strength of MPS in identifying a mutation in only one individual is emphasized in this study, where a de novo mutation was identified in a case of sporadic deafness. While it is not always possible to conclude that a mutation is pathogenic with only one individual, the evidence for p.Gln79* being the causative mutation for deafness in proband Z715 is strong. The c.853_854del mutation segregates in a family where the father, two daughters and a grandchild have the deletion, leading to profound deafness and cochlear malformation in the males, and mild hearing loss in the two carrier females, a phenomenon well documented in the literature (reviewed in [Marlin et al., 2009]). POU3F4 mutations are commonly linked with temporal bone abnormalities including Mondini malformation, a cochlea of one-and-one-half turns instead of the normal two-and-one-half turns and have been found in males, given the nature of the chromosome X linkage. To date, 13 deafness genes have been described in the Israeli Jewish population [Brownstein and Avraham, 2009; Brownstein et al., 2011; Horn et al., 2013]. Here, we identify an additional gene, POU3F4, with a current tally of 14 in the Israeli Jewish population. Further advances in sequencing chemistry and MPS data processing tools will help further speed up novel gene and mutation discoveries for deafness with the present ability to screen for all known deafness-causing genes in a single test [Shearer et al., 2011; Brownstein et al., 2012].
The mutations we have identified in human and mouse POU3F4 are predicted to lead to truncations of the protein. Pou3f4 is predicted to contain three nuclear localization signals (NLS), with one in the POUS and two in the POUHD [Lee et al., 2009]. In a study describing human mutations in POU3F4 in Korean DFNX2 families, transiently expressed mutant proteins of three mutations demonstrated a subcellular localization shift to the cytoplasm in a C3H10T1/2 cell line. One mutation, p.R329P, led to a substitution in the POUHD within an NLS motif, another caused a deletion in the POUHD in between two NLSs (p.S310del), and a third led to an early protein truncation that abolished both of the functional DNA binding domains and all three predicted NLSs (p.A116fs). In all three of these cases the expressed protein was confined primarily to the cytoplasm. Analysis of recently found POU3F4 mutations demonstrated similar findings for truncating mutations, with loss of transcriptional activity, as measured by cell transfections and corresponding luciferase assays [Choi et al., 2012]. In an Israeli family with progressive hearing loss, a truncated POU4F3 protein with a loss of the NLS led to a similar cytoplasmic mislocalization [Weiss et al., 2003]. Here we also demonstrate a similar effect with the human p.Ile285Argfs*43 and the mouse Pou3f4sdl p.Cys300* nonsense mutation transfected into COS-7 cells, and predicted to eliminate the third NLS of Pou3f4. Importantly, we demonstrate the mislocalization of mutant Pou3f4 for the first time in vivo in the mammalian inner ear. Immunohistochemical analysis of the sdl mouse mutant demonstrated that while the Pou3f4 mutant protein is present, it was mislocalized in the mesenchymal cells of the spiral ligament and spiral limbus, two tissues in which Pou3f4 is normally expressed. Therefore, despite its presence in mutant cells in its truncated form, its cytoplasmic mislocalization most likely reflects lack of functionality.
Based on the physiological expression patterns of Pou3f4 in the inner ear, early studies had focused on the otic mesenchyme. In recent years, it has become evident that the regulation of inner ear development and function by Pou3f4 is highly complex and is mediated through numerous downstream targets regulating diverse biological functions, including ion transport [Xia et al., 2002; Song et al., 2011], auditory spiral ganglion axon growth [Coate et al., 2012] and retinoic acid, Fgf and Hedgehog signaling pathways [Robert-Moreno et al., 2010]. Recently it was shown that Pou3f4 is capable of reprogramming fibroblasts into neural stem cells in combination with other transcription factors [Han et al., 2012]. Our study provides the first documentation of outer hair cell degeneration as a result of a Pou3f4 mutation. Outer hair cells do not express detectable amounts of Pou3f4 [Phippard et al., 1998] (Fig. 4C), but these cells are still subject to degradation and cell death in Pou3f4 mutants, and emphasizes the tight epithelial-mesenchymal interplay regulated by Pou3f4 in the inner ear.
In conclusion, the novel POU3F4 human and the mouse mutations elegantly complement each other, with the Pou3f4sdl mutant providing evidence of the mechanism leading to deafness. Further identification of downstream targets and deciphering of the protein network involved in Pou3f4 regulation will lead to a better understanding of the underlying molecular mechanisms leading to DFNX2 deafness. MPS, proven to be effective for detecting de novo mutations in sporadic cases, will continue to speed up the discovery of novel genes and mutations involved in inner ear pathologies, paving the way towards a more comprehensive understanding of pathways involved in auditory function.
Acknowledgments
We thank the families for their willingness to take part in our research study. We also thank Thomas Coate and Matthew Kelley for the Pou3f4 antibody and Vered Holdengreber, Elfi Holupirek, Auke Boersma, and Susan Marschall for technical assistance. This work has been funded by European Commission FP6 Integrated Project EUMODIC 037188 (M.H.A. and K.B.A.); the National Institutes of Health (NIDCD) R01DC011835, I-CORE Gene Regulation in Complex Human Disease Center No. 41/11, and the Hedrich Charitable Trust (K.B.A.); Israel Ministry of Science and Technology (D.R.L.); EMBO Molecular Medicine Fellowship AMMF 52-2010 (T.P.); the German Federal Ministry of Education and Research (NGFN-Plus grant No. 01GS0850 (M.H.A.) and 01GS0851 (E.W.); the Infrafrontier Grant 01KX1012 and German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD) (M.H.A.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Acevedo-Arozena A, Wells S, Potter P, Kelly M, Cox RD, Brown SD. ENU mutagenesis, a way forward to understand gene function. Annu Rev Genomics Hum Genet. 2008;9:49–69. doi: 10.1146/annurev.genom.9.081307.164224. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers CA, Lunter G, Macarthur DG, Mcvean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Bolado G, Rosenfeld MG, Swanson LW. Model of forebrain regionalization based on spatiotemporal patterns of POU-III homeobox gene expression, birthdates, and morphological features. J Comp Neurol. 1995;355:237–295. doi: 10.1002/cne.903550207. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Turnpenny P, Hoglund P, Kaariainen H, Sankila EM, van der Maarel SM, de Kok YJ, Ropers HH, Cremers FP, Pembrey M, et al. Further mutations in Brain 4 (POU3F4) clarify the phenotype in the X-linked deafness, DFN3. Hum Mol Genet. 1995;4:1467–1469. doi: 10.1093/hmg/4.8.1467. [DOI] [PubMed] [Google Scholar]
- Braunstein EM, Crenshaw EB, 3rd, Morrow BE, Adams JC. Cooperative function of Tbx1 and Brn4 in the periotic mesenchyme is necessary for cochlea formation. J Assoc Res Otolaryngol. 2008;9:33–43. doi: 10.1007/s10162-008-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Hardisty-Hughes RE, Mburu P. Quiet as a mouse: dissecting the molecular and genetic basis of hearing. Nat Rev Genet. 2008;9:277–290. doi: 10.1038/nrg2309. [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Avraham KB. Deafness genes in Israel: implications for diagnostics in the clinic. Pediatr Res. 2009;66:128–134. doi: 10.1203/PDR.0b013e3181aabd7f. [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol. 2012;13:245. doi: 10.1186/gb-2012-13-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, Mclellan MD, Larson DE, Kalicki JM, Pohl CS, Mcgrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BY, Kim DH, Chung T, Chang M, Kim EH, Kim AR, Seok J, Chang SO, Bok J, Kim D, Oh SH, Park WY. Destabilization and mislocalization of POU3F4 by C-terminal frameshift truncation and extension mutation. Hum Mutat. 2013;34:309–316. doi: 10.1002/humu.22232. [DOI] [PubMed] [Google Scholar]
- Coate TM, Raft S, Zhao X, Ryan AK, Crenshaw EB, 3rd, Kelley MW. Otic mesenchyme cells regulate spiral ganglion axon fasciculation through a Pou3f4/EphA4 signaling pathway. Neuron. 2012;73:49–63. doi: 10.1016/j.neuron.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers HH, Cremers FP. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- de Kok YJ, Vossenaar ER, Cremers CW, Dahl N, Laporte J, Hu LJ, Lacombe D, Fischel-Ghodsian N, Friedman RA, Parnes LS, Thorpe P, Bitner-Glindzicz M, et al. Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet. 1996;5:1229–1235. doi: 10.1093/hmg/5.9.1229. [DOI] [PubMed] [Google Scholar]
- Dror AA, Avraham KB. Hearing loss: Mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009;43:411–437. doi: 10.1146/annurev-genet-102108-134135. [DOI] [PubMed] [Google Scholar]
- Dror AA, Politi Y, Shahin H, Lenz DR, Dossena S, Nofziger C, Fuchs H, Hrabe de Angelis M, Paulmichl M, Weiner S, Avraham KB. Calcium oxalate stone formation in the inner ear as a result of an Slc26a4 mutation. J Biol Chem. 2010;285:21724–21735. doi: 10.1074/jbc.M110.120188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Tapia N, Hermann A, Hemmer K, Hoing S, Arauzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, Greber B, Yang JH, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Shalit E, Rzadzinska AK, Dror AA, Song L, Ron U, Tan JT, Shitrit AS, Fuchs H, Hasson T, Ben-Tal N, Sweeney HL, et al. A Myo6 mutation destroys coordination between the myosin heads, revealing new functions of myosin VI in the stereocilia of mammalian inner ear hair cells. PLoS Genet. 2008;4:e1000207. doi: 10.1371/journal.pgen.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, Friedman LM, Roux KJ, Kozlov S, Jeang K-T, Frydman M, Burke B, et al. The LINC complex is essential for hearing. J Clin Invest. 2013;123:740–750. doi: 10.1172/JCI66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- Huebner AK, Gandia M, Frommolt P, Maak A, Wicklein EM, Thiele H, Altmuller J, Wagner F, Vinuela A, Aguirre LA, Moreno F, Maier H, et al. Nonsense mutations in SMPX, encoding a protein responsive to physical force, result in X-chromosomal hearing loss. Am J Hum Genet. 2011;88:621–627. doi: 10.1016/j.ajhg.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter-Duvar IM. A technique for preparation of cochlear specimens for assessment with the scanning electron microscope. Acta Otolaryngol Suppl. 1978;351:3–23. doi: 10.3109/00016487809122718. [DOI] [PubMed] [Google Scholar]
- Hussain MA, Lee J, Miller CP, Habener JF. POU domain transcription factor brain 4 confers pancreatic alpha-cell-specific expression of the proglucagon gene through interaction with a novel proximal promoter G1 element. Mol Cell Biol. 1997;17:7186–7194. doi: 10.1128/mcb.17.12.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE. The paintfill method as a tool for analyzing the three-dimensional structure of the inner ear. Brain Res. 2006;1091:270–276. doi: 10.1016/j.brainres.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Song MH, Kang M, Lee JT, Kong KA, Choi SJ, Lee KY, Venselaar H, Vriend G, Lee WS, Park HJ, Kwon TK, et al. Clinical and molecular characterizations of novel POU3F4 mutations reveal that DFN3 is due to null function of POU3F4 protein. Physiol Genomics. 2009;39:195–201. doi: 10.1152/physiolgenomics.00100.2009. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Han D, Li J, Han B, Ouyang X, Cheng J, Li X, Jin Z, Wang Y, Bitner-Glindzicz M, Kong X, Xu H, et al. Loss-of-function mutations in the PRPS1 gene cause a type of nonsyndromic X-linked sensorineural deafness, DFN2. Am J Hum Genet. 2010;86:65–71. doi: 10.1016/j.ajhg.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin S, Moizard MP, David A, Chaissang N, Raynaud M, Jonard L, Feldmann D, Loundon N, Denoyelle F, Toutain A. Phenotype and genotype in females with POU3F4 mutations. Clin Genet. 2009;76:558–563. doi: 10.1111/j.1399-0004.2009.01215.x. [DOI] [PubMed] [Google Scholar]
- Minowa O, Ikeda K, Sugitani Y, Oshima T, Nakai S, Katori Y, Suzuki M, Furukawa M, Kawase T, Zheng Y, Ogura M, Asada Y, et al. Altered cochlear fibrocytes in a mouse model of DFN3 nonsyndromic deafness. Science. 1999;285:1408–1411. doi: 10.1126/science.285.5432.1408. [DOI] [PubMed] [Google Scholar]
- Nance WE. The genetics of deafness. Ment Retard Dev Disabil Res Rev. 2003;9:109–119. doi: 10.1002/mrdd.10067. [DOI] [PubMed] [Google Scholar]
- Petersen MB, Wang Q, Willems PJ. Sex-linked deafness. Clin Genet. 2008;73:14–23. doi: 10.1111/j.1399-0004.2007.00913.x. [DOI] [PubMed] [Google Scholar]
- Petit C, Richardson GP. Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci. 2009;12:703–710. doi: 10.1038/nn.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K, Luisi B. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol. 2000;302:1023–1039. doi: 10.1006/jmbi.2000.4107. [DOI] [PubMed] [Google Scholar]
- Phippard D, Heydemann A, Lechner M, Lu L, Lee D, Kyin T, Crenshaw EB., 3rd Changes in the subcellular localization of the Brn4 gene product precede mesenchymal remodeling of the otic capsule. Hear Res. 1998;120:77–85. doi: 10.1016/s0378-5955(98)00059-8. [DOI] [PubMed] [Google Scholar]
- Phippard D, Lu L, Lee D, Saunders JC, Crenshaw EB., 3rd Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. J Neurosci. 1999;19:5980–5989. doi: 10.1523/JNEUROSCI.19-14-05980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippard D, Boyd Y, Reed V, Fisher G, Masson WK, Evans EP, Saunders JC, Crenshaw EB., 3rd The sex-linked fidget mutation abolishes Brn4/Pou3f4 gene expression in the embryonic inner ear. Hum Mol Genet. 2000;9:79–85. doi: 10.1093/hmg/9.1.79. [DOI] [PubMed] [Google Scholar]
- Reardon W. Sex linked deafness: Wilde revisited. J Med Genet. 1990;27:376–379. doi: 10.1136/jmg.27.6.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A, Naranjo S, de la Calle-Mustienes E, Gomez-Skarmeta JL, Alsina B. Characterization of new otic enhancers of the pou3f4 gene reveal distinct signaling pathway regulation and spatio-temporal patterns. PLoS One. 2010;5:e15907. doi: 10.1371/journal.pone.0015907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, Haas SA, Weegerink NJ, Oostrik J, Hu H, Hoefsloot LH, Kannan S, Huygen PL, Pennings RJ, Admiraal RJ, Kalscheuer VM, Kunst HP, et al. Next-generation sequencing identifi es mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am J Hum Genet. 2011;88:628–634. doi: 10.1016/j.ajhg.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Hildebrand MS, Sloan CM, Smith RJ. Deafness in the genomics era. Hear Res. 2011;282:1–9. doi: 10.1016/j.heares.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MH, Choi SY, Wu L, Oh SK, Lee HK, Lee DJ, Shim DB, Choi JY, Kim UK, Bok J. Pou3f4 deficiency causes defects in otic fibrocytes and stria vascularis by different mechanisms. Biochem Biophys Res Commun. 2011;404:528–533. doi: 10.1016/j.bbrc.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Van Camp G, Smith RJH. [Accessed March 2013];Hereditary Hearing Loss Homepage. 2013 Available at: http://hereditaryhearingloss.org.
- Weiss S, Gottfried I, Mayrose I, Khare SL, Xiang M, Dawson SJ, Avraham KB. The DFNA15 deafness mutation affects POU4F3 protein stability, localization, and transcriptional activity. Mol Cell Biol. 2003;23:7957–7964. doi: 10.1128/MCB.23.22.7957-7964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia AP, Kikuchi T, Minowa O, Katori Y, Oshima T, Noda T, Ikeda K. Late-onset hearing loss in a mouse model of DFN3 non-syndromic deafness: morphologic and immunohistochemical analyses. Hear Res. 2002;166:150–158. doi: 10.1016/s0378-5955(02)00309-x. [DOI] [PubMed] [Google Scholar]