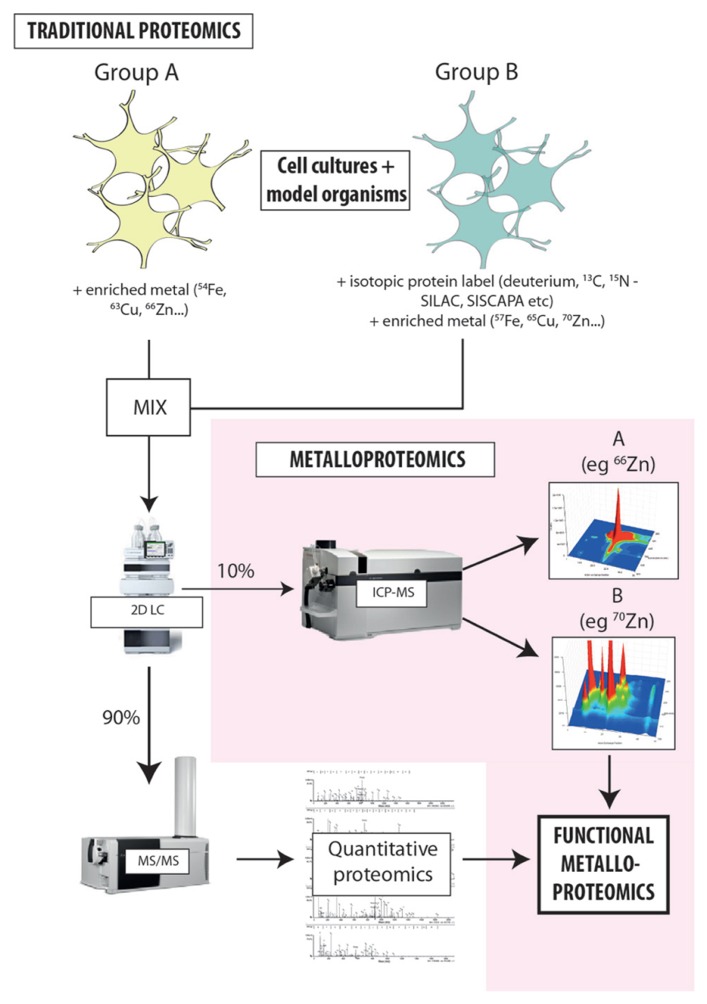
FIGURE 2.
Proposed workflow for integrated metalloproteomics. Hyphenating native separation techniques to ICP-MS is the key to unlocking the secrets of metals and protein function. Rather than relying solely on bulk measures, directly associating metals with specific proteins provides new insight into how metals carry out biochemical processes in the cell. At the current state of our knowledge about the level of metalloproteins in biology the coupling of size exclusion to ICP-MS have the promise of being quantitative thus allowing the comparison of different samples and detail about the global or metalloprotein specific changes that occur. Despite SEC being a low resolution technique it can allow researchers to make educated guesses about the ID of proteins of interests based on their MW. Continued evolution of hyphenated LC-ICP-MS will only increase the arsenal of tools at our disposal to discover, identify, and characterize metalloproteins. The workflow developed in our laboratory adapts existing isotope labeling techniques used for proteomics [such as SILAC (stable isotope labeling by amino acids in cell culture) and SISCAPA (stable isotope standards and capture by anti-peptide antibodies)] to include the addition of isotopically enriched metal salts, providing new opportunities to probe the direct relationship metal cofactors have with protein function and allowing simultaneous analysis of both metals and proteins in individual experimental groups. Highly sensitive, isotope-specific ICP-MS detection is used to align metal distribution with quantitative proteomics, directly associating the presence of a protein species with a specific, metal-mediated function. This approach is extremely cost-effective, and can be seamlessly integrated into existing workflows with minimal disruption to the standard laboratory process.