Figure 1.
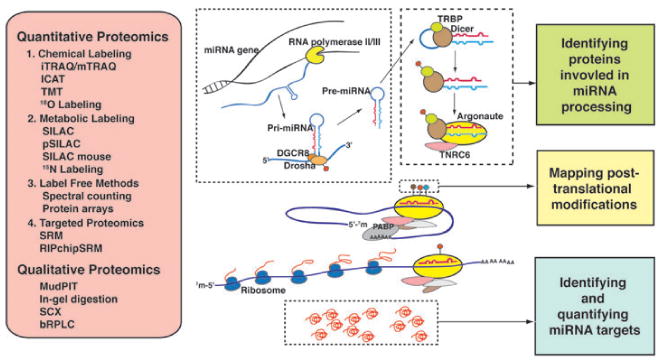
Proteomic strategies for studying the mechanism of miRNA processing and miRNA-mediated translation repression. Genes encoding miRNAs are transcribed as pri-miRNAs by RNA polymerase II and processed by the microprocessor complex consisting of Drosha and DGCR8. This complex cleaves pri-miRNAs into ∼60–80 nt hairpin structures, called pre-miRNAs, which are transported out of nucleus by exportin-5. In the cytoplasm, Dicer associates with TRBP and excises the loop portion of pre-miRNAs generating double stranded mature miRNAs. These are loaded onto RNA interference silencing complex (RISC) consisting of Argonaute and TNRC6. Translation inhibition is brought about by base-pairing of guide strand miRNA to the 3′UTR of mRNA and interfering with translation initiation. Quantitative proteomics analyses encompass different categories of strategies including chemical labeling, metabolic labeling, label-free methods and targeted proteomics. Among them, iTRAQ and SILAC are the most frequently used. Along with various techniques in qualitative proteomics, these methods can enable not only characterization of the miRNA processing pathway components and their posttranslational modifications but also discovery of miRNA targets.
