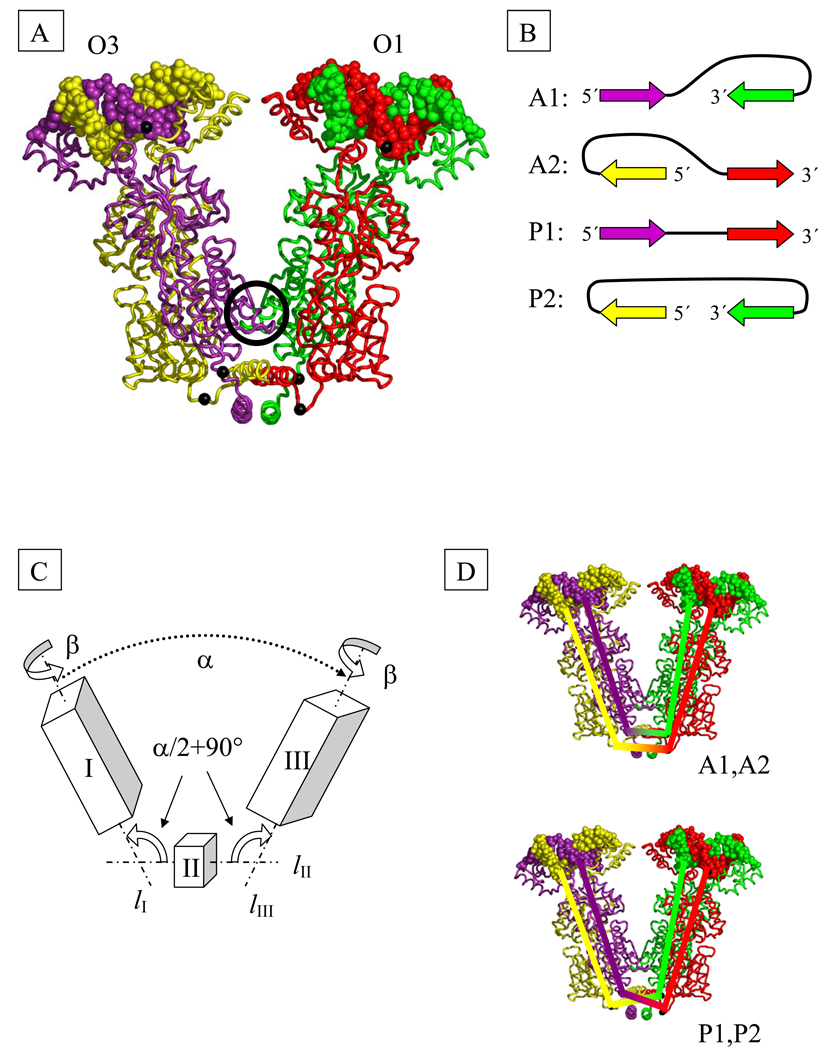
Fig. 2.
(A) Model of the structure of the tetrameric Lac repressor protein (LacR) in complex with O1 and O3 operator segments, obtained by composition of available X-ray data (see Methods). The black spheres on protein represent the Cα atoms of Gln 335 and those on DNA the P atoms of the central base pairs. Color-coding denotes the protein monomers and DNA chains in closest contact at the highlighted P atoms. The black circle marks the dimer contact interface found in the crystal structure.
(B) DNA loop types. The color-coded arrows depict the 5′-3′ directions of the sequence strand on LacR in the four possible orientations of DNA on the tetramer. The colors correspond to those of the associated DNA and protein chains in part (A).
(C) Schematic representation of LacR opening. The rigid domains I (residues 1–332 of chains A and B and the DNA bound to these chains) and III (corresponding residues of chains C and D and the bound DNA) are connected to domain II (residues 340–354 of chains A, B, C, D) by two hinges. The axes of rotational symmetry of the three domains are lI, lII, and lIII. Chains (A–D) correspond respectively to proteins shown in (A) in violet, yellow, green, and red.
(D) Schematic representation of the closures of DNA strands used in the computation of linking number. The top closure is appropriate for antiparallel loops and the bottom for parallel and extended loops.