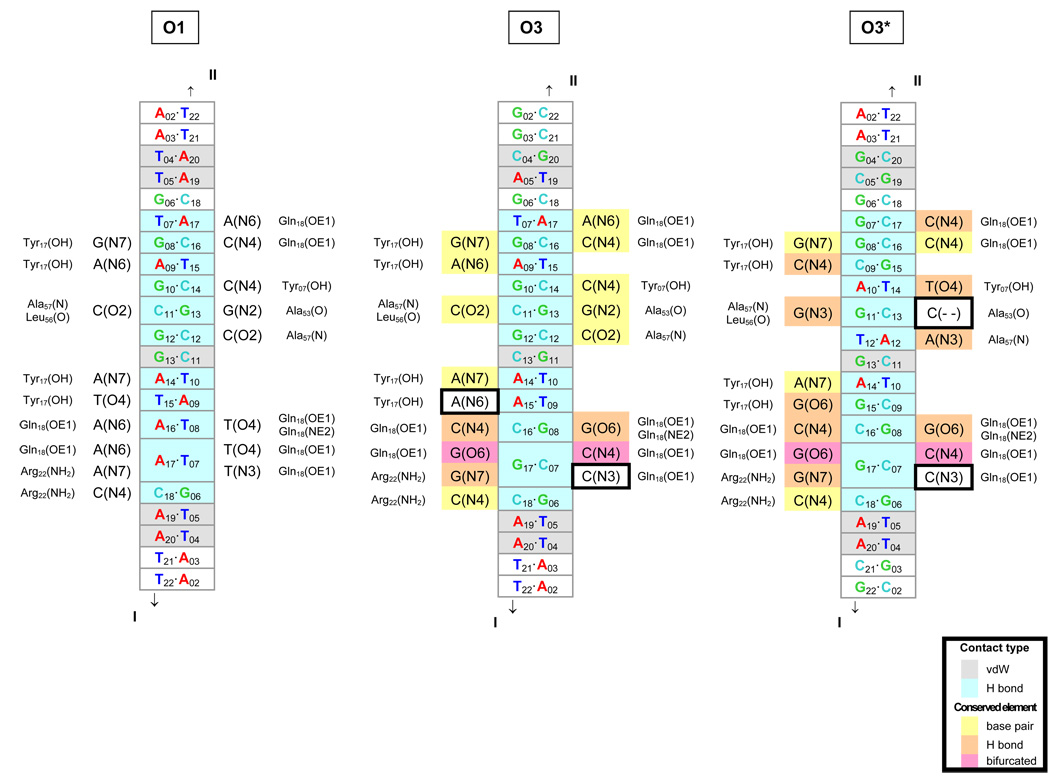
Fig. 5.
Diagrams of comparative molecular interactions of the LacR headpiece with the O1, O3, and O3* operators. Strand I is the leading strand of each recognition site, i.e., the sequences listed in Table 1–Table 2 for O1 and O3 and in Table 3 for O3*, and Strand II is the complement. The O1 interactions are as found in solution [10]; the O3 and O3* interactions are putative. DNA base pairs in close contact (interatomic distances of 3.4 Å or less) with protein residues in the O1 complex are denoted in gray (van der Waals’ interactions) and aqua (hydrogen bonds) in the center of each diagram. The contacted atoms of the bases are indicated on each side of the center column, together with the corresponding protein residues on the outside. The base pair contacts in O3 and O3* that are identical to those in O1 are highlighted in yellow, and the contacts that preserve hydrogen bonding are denoted in orange. The bifurcated hydrogen bonding (at position 17) that accommodates all base-pair combinations is shown in pink. Comparison of the three sites shows that most of the hydrogen-bonding elements are conserved upon substitution of the O3 or O3* sequence—a direct consequence of the isosteric character of the Watson-Crick base pairs, the pseudo-symmetric positioning of hydrogen-bond donor atoms in the DNA minor groove [48], the equivalent positioning of major-groove atoms in conservatively substituted (G↔T and A↔C) bases [48], and the dual hydrogen-bonding (donor or acceptor) capabilities of selected amino acids. The two potential hydrogen-bonding elements not conserved upon substitution of the O3 or O3* sequences are outlined by boxes.