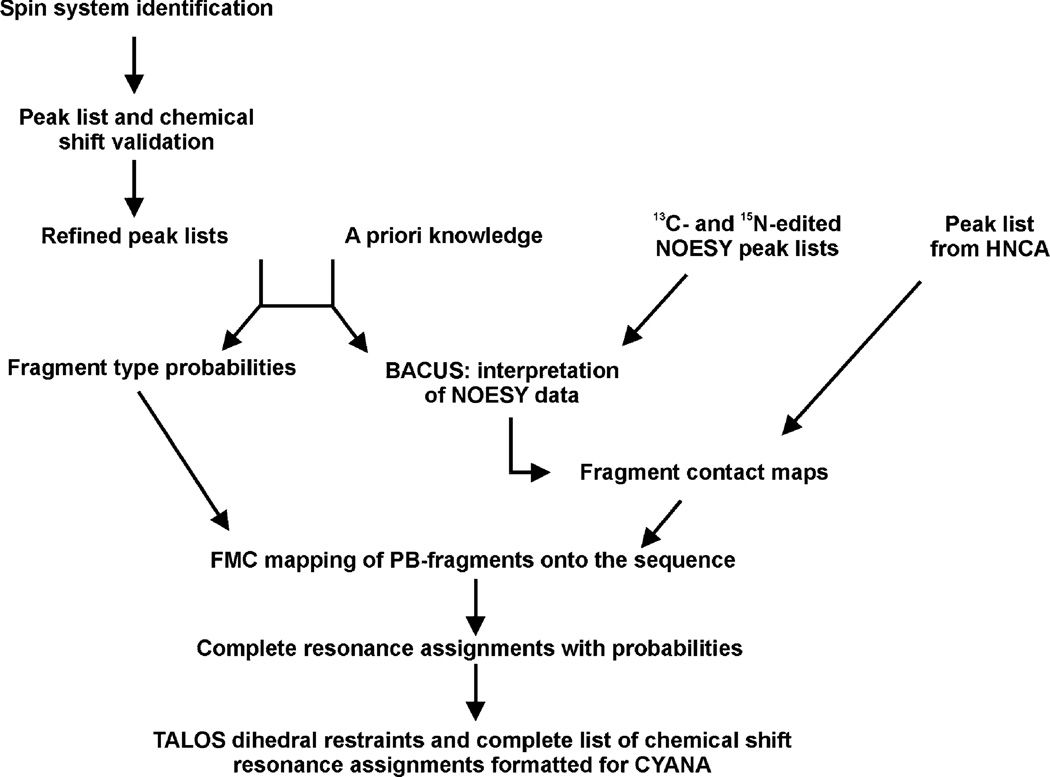
Fig. 3.
Overview of ABACUS and structure calculation workflow. Peaks are manually picked in the HNCO, HNCA, CBCA(CO)NH, HBHA (CBCACO)NH, H(C)CH- and (H)CCH-TOCSY experiments, along with those obtained from the HNCA and 13C- and 15N-edited NOESY. These peak lists are prerequisites for the ABACUS protocol. A reference BMRB chemical shift list identifies potential mismatches or deviations between experimental spin systems and those of standard amino acids. The amino acid sequence and the peak lists from the scalar-coupled experiments are used to match individual spin systems with corresponding amino acid types. NOESY and HNCA data are used to establish connectivities (e.g. contact maps). Fragment type probabilities and contact maps are utilized in FMC simulations to map corresponding spin systems to specific positions in the amino acid sequence. The result is a complete chemical shift list that is used to generate angular and distance constraints for subsequent use in CYANA