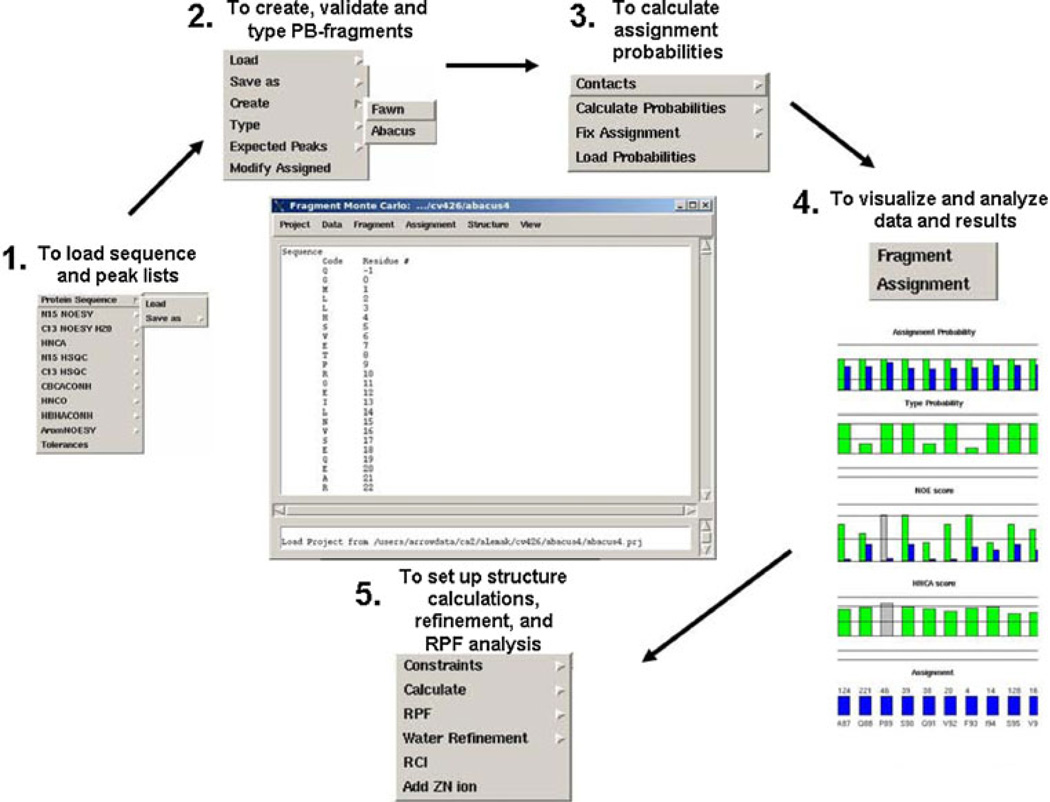
Fig. 5.
FMCGUI overview. The process begins by loading in the amino acid sequence and respective peak lists from corresponding experiments defined in the minimal dataset (Panel 1). Tolerances are set to improve the reliability of the automated assignment process. The Fragment menu item helps to organize the information for each PB spin system into individual fragments (Panel 2). Once all of the fragments (spin systems) are assembled, FMCGUI has a built-in chemical shift databank that quickly searches for errors in the peak picking process. In addition, the user-friendly interface identifies potential errors in the peak lists which may produce errors in the assignment list. Assignment probabilities are determined using a Fragment Monte Carlo routine with ABACUS and/or FAWN approaches (Panel 3). Confidence in the final chemical shift assignment list is highly dependent on the NOE and HNCA scores (Panel 4). These data are easily manipulated and can be readily exported into CYANA and TALOS formats (Panel 5). Structural ensembles are read back into FMCGUI to assist in the calculation of recall and precision scores and structure refinement in CNS