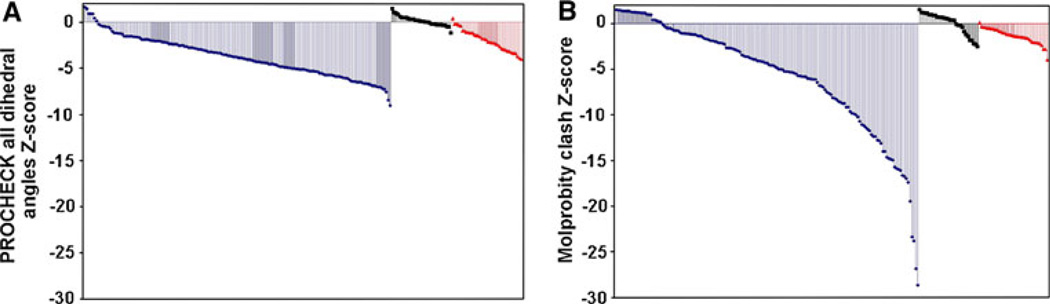
Fig. 6.
Approach yields high quality protein structures. As one measure of structure quality, we compare the PROCHECK all dihedral angle (a) and Molprobity clash Z-scores (b) for NUS/MDD/ ABACUS derived structures (red; PDB accession codes 2JTV, 2KP6, 2KEO, 2KFV, 2KQ9, 2K4X, 2JYN, 2×8N, 2K8E, 2K2P, 2K28, 2JOQ, 2KKX, 2KR1, 2KO6, 2KKY, 2JXX, 2JQ4, 2JYA, 2K4V, 2KDB, 2JQ5, 2K54, 2K1B, 2KLC, 2K7I, 2KNR, 2KCO, 2K2C, 2KGO, 2IDA, 2KVR, 2JUF, 2KJZ, 2JN4, 2KKU), high-resolution X-ray crystal structures of similar sized proteins deposited in April 2009 with a resolution of < 2Å (black), and other similarly sized proteins determined by conventional NMR methods from non-Structural Genomics groups (blue), deposited in the PDB from January 1st, 2008–June 30th, 2009