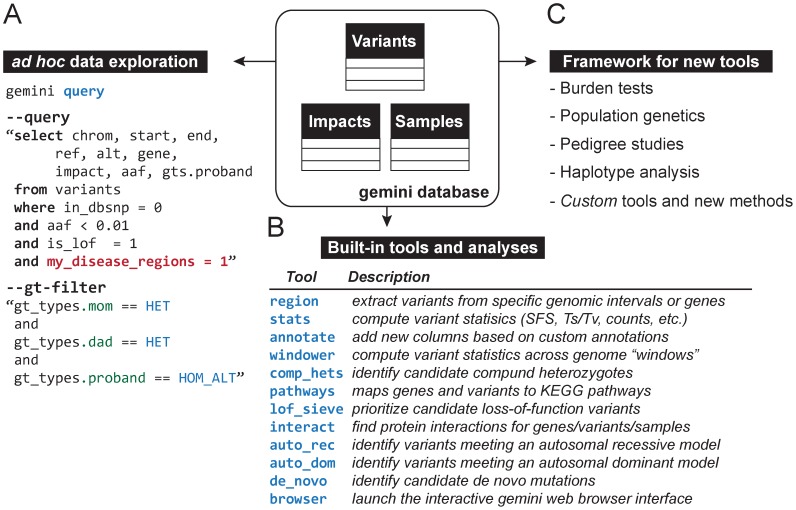
Figure 2. Variant mining and tool development with the GEMINI database framework.
(A) Storing variants and annotations in the same database framework enables ad hoc SQL data exploration through both the query module and a Python programming interface. Analysis queries can filter variants based on pre-installed annotations (e.g., in_dbsnp = 0) and custom annotations (e.g., my_disease_regions = 1). Users may also select and filter variants based upon the genotypes of specific individuals (e.g., gt_types.mom = = HET), thus allowing one to identify variants meeting specific inheritance patters, as shown here. (B) The GEMINI database framework also enables the development of tools that facilitate automated analyses for routine analysis tasks. (C) Moreover, it serves as a standard interface for developers to develop new tools and algorithms and to implement improved statistical tests for population and medical genetics.