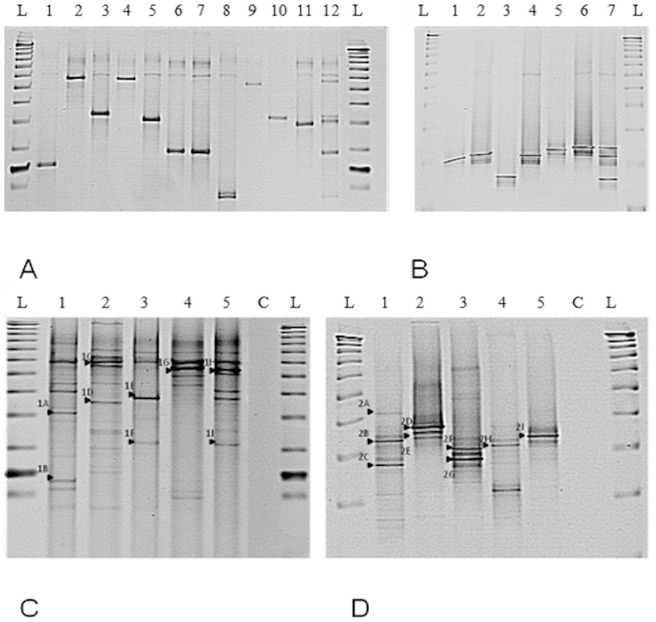
Figure 1. DGGE EM profiling using 16S rRNA gene PCR amplicons.
(A) Profiles of individual EM type strains obtained using the Mycobacterium genus specific primer set JSY16S. L is the reference ladder (TrackIt™ 50 bp DNA Ladder, Invitrogen, Ltd., Paisley, UK), lanes 1–12 are, respectively: M. smegmatis, M. aichiense, M. aurum, M. gilvum, M. phlei, M. agri, M. peregrinum, M. duvalii, M. abscessus, M. fortuitum, M. vaccaeand a mixture of equimolar quantities of the above listed EM species. (B) Profiles obtained using the slow mycobacteria specific primer set APTK16S. L is a reference ladder (TrackIt™ 50 bp DNA Ladder, Invitrogen, Ltd., Paisley, UK), lanes 1–7 are, respectively: M. intracellulare, M. marinum, M. kansasii, M. xenopi, M. aviumparatuberculosis, M. bovis BCG and a mixture of equimolar quantities of the above listed EM species. (C) EM soil community profiling using the Mycobacterium genus specific primers (JSY16S). L is a reference sizing ladder (TrackIt™ 50 bp DNA Ladder, Invitrogen, Ltd., Paisley, UK), C is the negative PCR control, samples 1 – 4 are the four Ethiopian soils (1108, 1109,1110, 1111) and 5 is Cryfield. The arrows (1A–1I) indicate the bands that were excised and sequenced (Table 4). (D) EM soil community profiling using the slow grower mycobacteria specific 16S rRNA gene specific primers (APTK16S). L is a reference sizing ladder (TrackIt™ 50 bp DNA Ladder, Invitrogen, Ltd., Paisley, UK), C is the negative PCR control, samples 1–4 are the four Ethiopian soils (1108, 1109, 1110, 1111) and 5 is Cryfield. The arrows (2A–2I) represent the bands that were excised and sequenced (Table 4).