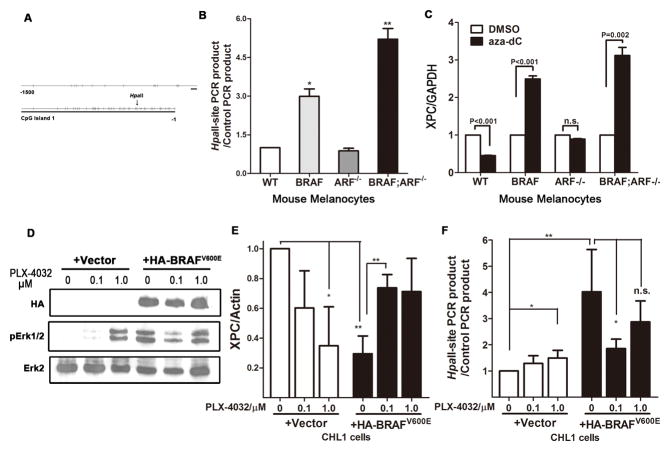
Figure 6. Mutant BRAFV600E caused XPC promoter methylation.
(A) Diagram showing CpG islands in promoter region of the mouse XPC gene.
(B) Detection of XPC promoter methylation in mouse primary melanocytes of different genotypes. Genomic DNA was subjected to methylation-sensitive HpaII treatment, following by quantitative PCR using primers flanking the HpaII-recognition sites. Primers flanking genomic sequences that do not contain any HpaII site, thereby resistant to HpaII treatment, were used as internal control. Increased recovery of PCR product indicates increased methylation in the corresponding region. Student t-test, * p<0.05, ** p<0.005.
(C) Expression of XPC following treatment with the demethylating agent 5′-aza-2′-deoxycytidine (aza-dC). Mouse primary melanocytes of different genotypes were treated with 5μM aza-dC for 48h, followed by RNA preparation and quantitative real-time PCR.
(D–E) Acute expression of BRAFV600E increases XPC promoter methylation and inhibits XPC mRNA expression. BRAF wild-type, NRAS wild-type CHL1 melanoma cells were infected with either empty vector or HA-BRAFV600E virus, followed by treatment with mutant BRAF inhibitor PLX-4032 at indicated concentration for 48h. Protein, RNA and DNA were collected and subjected to immunoblot to confirm the activation of BRAF pathway (D), real-time quantitative PCR to determine XPC mRNA level (E) and HpaII-based qPCR to examine promoter methylation (F).