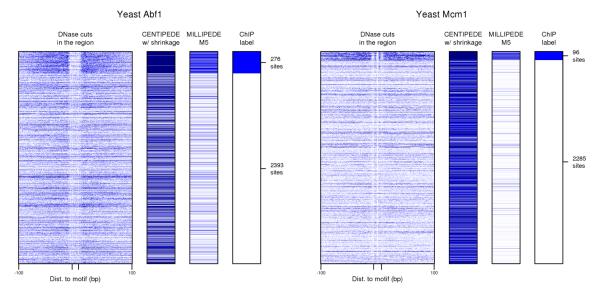
Fig. 6.
DNase data can exhibit systematic artifacts such as sequence-dependent digestion bias. Left and right panels show yeast Abf1 and Mcm1 candidate binding sites, respectively. Notice that some fine details of the DNase cut data are preserved within and around candidate binding sites, whether the site is bound or unbound. centipede is prone to over-fit these details both because of the large number of parameters in the multinomial component of its likelihood, and because it is unsupervised and uses EM to assign labels to candidate sites.