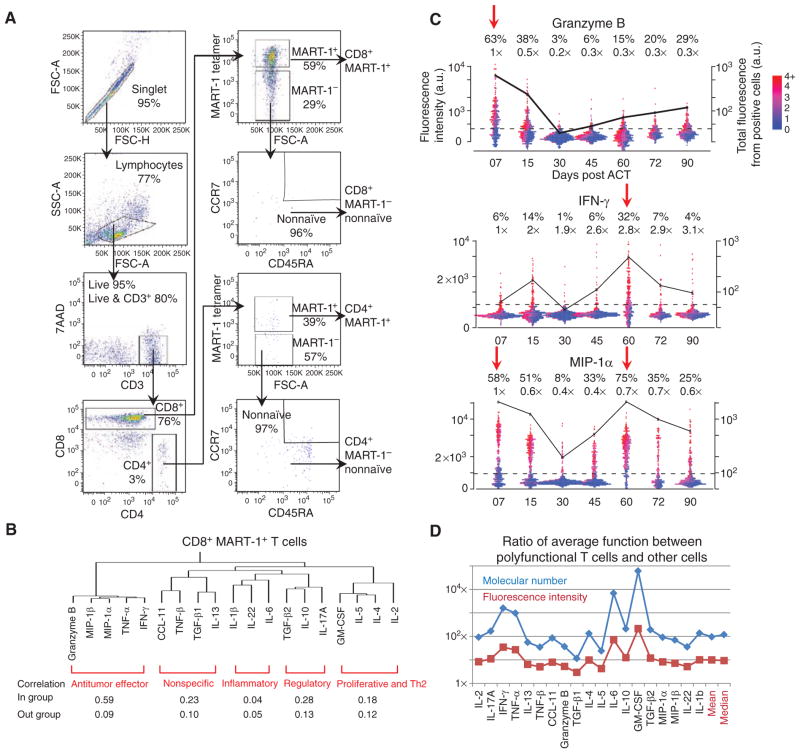
Figure 2.
FACS scheme for purifying phenotypically defined T cells and general properties of T-cell functions. A, multiparametric FACS purification for selecting phenotypically defined T-cell populations. A representative set of scatterplots is shown with the surface markers used and the cell frequency for each gating. B, hierarchical clustering of the 19 functional cytokines studied on the basis of the single-cell cytokine secretion measurement of CD8+ MART-1+ T cells from all 3 patients, and across all time points. Each functional group is identified and labeled (red). Protein–protein correlations for proteins within the same group and across groups are given below the clustering map. C, one-dimensional scatterplots of 3 representative cytokines produced by single cells, separated by time points. The dotted line represents the gate that separates cytokine-producing and nonproducing cells. The percentages given above the plots denote the frequency of positive cells and the relative mean fluorescence intensity (MFI) of those cells relative to day 7. Each point represents a single-cell assay. The points are color encoded (from purple to red) to represent the number of different proteins produced by each cell. The black trend line shows the total functional intensity of the positive cells for the specific cytokine plotted, computed as the frequency of positive cells, multiplied by their MFI. D, ratio between the MFI (red line) and the molecular number of cytokines (blue line) for polyfunctional T cells (cells with 5 or more functions) and all other cells, for each cytokine. At far right are the mean and median values, averaged over all cytokines. 7-AAD, 7-aminoactinomycin D; CCL, CC chemokine ligand; FSC, forward scatter; GM-CSF, granulocyte macrophage colony-stimulating factor; MIP, macrophage-inflammatory protein; SSC, side scatter.