Figure 3.
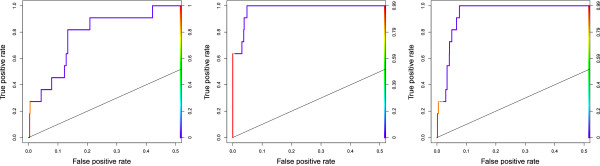
E. coli: RcsB motif ROC curves. ROC curves (plotted for 0≤sFPR≤0.5) for the most likely E.coli RcsB motif, as chosen using the known width (left), MCOIN (centre) and E-values based estimator (right). The curve colour illustrates the threshold of p(Zi,j = 1|Xi,j,θ), from 1.0 (red) to 0.0 (blue). Although MCOIN and the E-values estimator both underestimate the known motif width, site-level predictions are improved as the true motif is relatively weakly discovered at the true width. Performance in terms of AUC may be increased by choosing stronger and/or unshifted motif models at non-optimal widths. MCOIN displays improvement over the known motif width and the E-values estimator in all three classification performance measures.
