Figure 8.
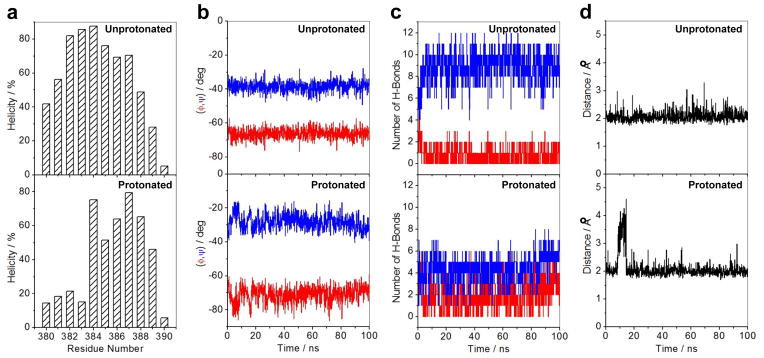
Effect of protonation on the structural stability of αII helix (residues 380–390) located within the ZFII of DB domain of EGR1 as determined from MD analysis on the structural models of the DB_WT domain of EGR1 containing H382 in unprotonated and protonated forms in complex with the ZRE duplex. Note that in the unprotonated form, H382 is protonated only at Nε2 atom within the imidazole ring, while it is protonated at both Nε2 and Nδ1 atoms in the protonated form. (a) Helicity as a function of residue number within the αII helix of ZFII in unprotonated (top panel) and protonated (bottom panel) forms of DB domain. Note that helicity is defined as the percentage of time each residue samples the α-helical conformation over the course of the simulation. (b) Dependence of backbone torsion angles φ (red) and ψ (blue) within the αII helix of ZFII in unprotonated (top panel) and protonated (bottom panel) forms of DB domain. (c) Variation of number of intramolecular backbone hydrogen bonds (H-bonds) between residue i and (i+3) characteristic of an 310-helical conformation (red) as well as between residue i and (i+4) characteristic of an α-helical conformation (blue) observed within the αII helix of ZFII in unprotonated (top panel) and protonated (bottom panel) forms of DB domain. (d) Distance between Hε2 atom (the hydrogen atom directly attached to Nε2) within the imidazole ring of H382 and N7 atom within the guanine base of G0 as a function of time in unprotonated (top panel) and protonated (bottom panel) forms of DB domain. Note that G0, according to the nomenclature shown in Figure 1b, is the central guanine of the middle trinucleotide subsite that accommodates ZFII within the DB domain.
