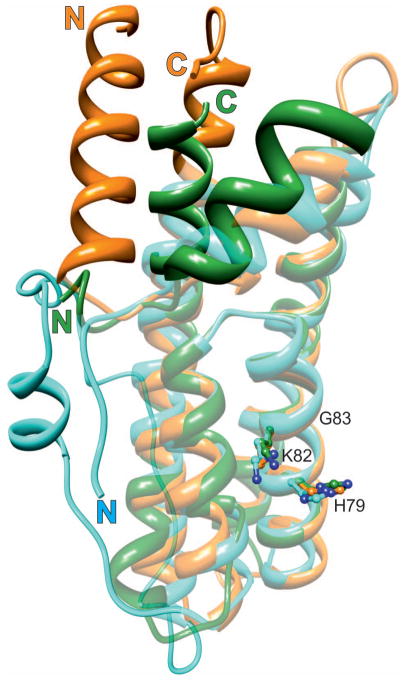
Fig. 5.
Structural alignment of HPt orthologs from three different kingdoms. MtHPt1 (plants), orange (this work); YDP1 (fungi), cyan (S. cerevisiae), and ArcB (bacteria), green (E. coli). The latter two proteins have been extracted from their complexes (PDB codes 1oxk and 1bdj, respectively). The well-superposed elements are shown in semi-transparent mode, the divergent elements are in solid color. Note the absence of helix α1 in the lower organisms and a long insert between helices α5 and α6 in YDP1. The active histidine (ball-and-stick model) occupies nearly the same position in all proteins. The lysine and glycine residues from the KGSS sequence motif that is conserved in all kingdoms are also shown to indicate the structural location of this motif in HPt proteins. These residues form part of the positively charged surface area (compare Fig. 6B). In general, the structural divergence increases with the distance from the active site. Numbering of the residues corresponds to MtHPt1.