Abstract
Background
Ribosome assembly cofactor RimP is one of the auxiliary proteins required for maturation of the 30S subunit in Escherichia coli. Although RimP in protein synthesis is important, its role in secondary metabolites biosynthesis has not been reported so far. Considering the close relationship between protein synthesis and the production of secondary metabolites, the function of ribosome assembly cofactor RimP on antibiotics production was studied in Streptomyces coelicolor and Streptomyces venezuelae.
Results
In this study, the rimP homologue rimP-SC was identified and cloned from Streptomyces coelicolor. Disruption of rimP-SC led to enhanced production of actinorhodin and calcium-dependent antibiotics by promoting the transcription of actII-ORF4 and cdaR. Further experiments demonstrated that MetK was one of the reasons for the increment of antibiotics production. In addition, rimP-SC disruption mutant could be used as a host to produce more peptidyl nucleoside antibiotics (polyoxin or nikkomycin) than the wild-type strain. Likewise, disruption of rimP-SV of Streptomyces venezuelae also significantly stimulated jadomycin production, suggesting that enhanced antibiotics production might be widespread in many other Streptomyces species.
Conclusion
These results established an important relationship between ribosome assembly cofactor and secondary metabolites biosynthesis and provided an approach for yield improvement of secondary metabolites in Streptomyces.
Keywords: rimP-SC, Streptomyces coelicolor, Actinorhodin, Calcium-dependent antibiotics
Introduction
In bacteria, more than 90% of energy is used in protein synthesis [1]. A large amount of them is used in ribosome assembly and protein translation. In vitro experiments have revealed that 50S and 30S ribosomal subunits could be reconstituted into active ribosomes from isolated components through heat-activation steps under different magnesium concentrations. However, these steps are not required and auxiliary proteins are needed in vivo[2]. An increasing number of ribosome assembly factors have been identified for 30S subunit reconstitution, such as RimP, RimM and RbfA in Escherichia coli[2].
RimP, formerly known as YhbC or P15a, is encoded by rimP in the rbfA operon and required for the maturation of 30S subunit. RimP is associated with 30S subunit but not 50S subunit or 70S ribosome. In the rimP deletion mutant, immature 16S rRNA is accumulated and the ribosomal profile shows fewer polysomes and the accumulation of unassociated 30S and 50S subunits. The difference becomes more obvious with the increasing temperature. The slow growth of rimP deletion mutant could not be suppressed by the increased expression of other known 30S maturation factors [2]. In vitro assembly studies showed that the preincubation of RimP with 16S rRNA could accelerate the binding rates of the 5′ domain ribosomal proteins S5 and S12 to almost all of the 3′ domain proteins (S3, S7, S9, S10, S13, and S14) [3,4].
Streptomyces coelicolor is the genetically most studied streptomycete and used as a model strain for studying the biology of actinomycetes [5,6]. It produces at least four distinct classes of antibiotics [6], including the well-known blue-pigmented aromatic polyketide antibiotic actinorhodin (ACT) which provides an easily tractable system for the methodological study of strain improvement [7], the red oligopyrrole prodiginine antibiotics (RED) [8], the acidic lipopeptide calcium-dependent antibiotics (CDA) [9] and methylenomycin [10]. The complete sequence and annotation of the S. coelicolor genome provide a way for its rational manipulation to identify potentially novel pathway products, and 29 predicted secondary metabolic gene clusters have been identified so far [11,12]. Besides screening new compounds, improving the production of existing compounds is still an important object, especially for the clinically and agriculturally applied antibiotics. Current main methods of improving antibiotics production include classical random mutation and ribosome engineering by the introduction of ribosomal protein mutations conferring drug resistance [13,14]. Although random mutation has played an important role in industry, its random nature is main drawback. In contrast, ribosome engineering approach allows for more rational manipulation.
In this paper, we cloned a rimP homologous gene rimP-SC from S. coelicolor and disruption of rimP-SC significantly increased the production of ACT and CDA. Meanwhile, the rimP-SC disruption mutant used as a heterologous expression host could produce more polyoxin or nikkomycin than the wild-type strain. In addition, disruption of rimP-SV also markedly improved jadomycin production in Streptomyces venezuelae, indicating that disruption of rimP homologues might be a widespread method for improving antibiotics production in Streptomyces.
Results
Identification of rimP homologue in S. coelicolor
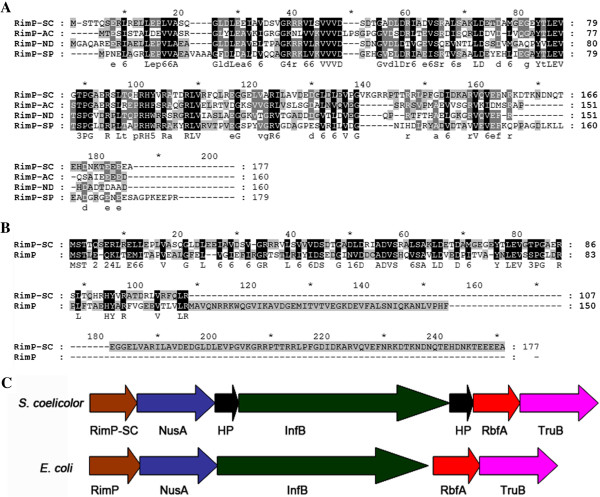
SCO5703 encodes a hypothetical protein consisting of 177 amino acids with a predicted molecular mass of 19.6 kDa. Comparative analysis demonstrated that its amino acid sequence was relatively conserved in actinomycetes (Figure 1A). However, its functional analysis has not been reported so far. Sequence alignment showed that it has 27% identity (41% similarity) with the RimP from E. coli (Figure 1B). Gene organization demonstrates that SCO5703 is flanked by genes similar to those found in E. coli, including the transcription elongation factor gene nusA, the translation initiation factor gene infB, the ribosome binding factor gene rbfA and the tRNA pseudouridine synthase gene truB (Figure 1C). The deduced product of SCO5703 contains the eukaryotic Sm or Sm-like (LSm) domain which associates with RNA to form the core domain of the ribonucleoprotein particle involved in a variety of RNA processing events including pre-mRNA splicing, telomere replication and mRNA degradation, making it a likely target for binding with the 30S ribosomal subunit in S. coelicolor.
Figure 1.
Alignment of RimP from different bacteria and genetic organization of the rimP-nusA-infB operon. (A) Alignment of RimP-SC with RimP-AC, RimP-ND and RimP-SP. (B) Alignment of RimP-SC with RimP. (C) Genetic organization of the rimP-nusA-infB operon in S. coelicolor and E. coli. RimP, ribosome maturation factor of E. coli; RimP-SC, RimP homologue of S. coelicolor; RimP-AC, RimP homologue of Actinomyces sp. oral taxon 848 str. F0332; RimP-ND, RimP homologue of Nocardiopsis dassonvillei; RimP-SP, RimP homologue of Saccharomonospora paurometabolica; NusA, the transcriptional elongation factor; InfB, the translational initiation factor; RbfA, ribosome binding factor; TruB, the tRNA (Ψ55) synthase; HP, hypothetical protein.
To study the function of SCO5703, E. coli rimP disruption mutant (rimPDM) was constructed by PCR-targeting strategy. Then, the heterologous complemented strain of rimPDM (rimPDMC) was also constructed. Finally, the growth rates of E. coli wild-type strain BW25113, rimPDM and rimPDMC were detected at 28°C, 37°C or 42°C. As reported previously [2], rimPDM showed a reduced growth rate, especially at higher temperature. Introduction of the intact SCO5703 into the rimPDM restored the slow-growth phenotype almost to the wild-type level, indicating that SCO5703 is a functional homolog of E. coli rimP and thus is designated as rimP-SC (data not shown).
Disruption of rimP-SC enhances antibiotics production in Streptomyces
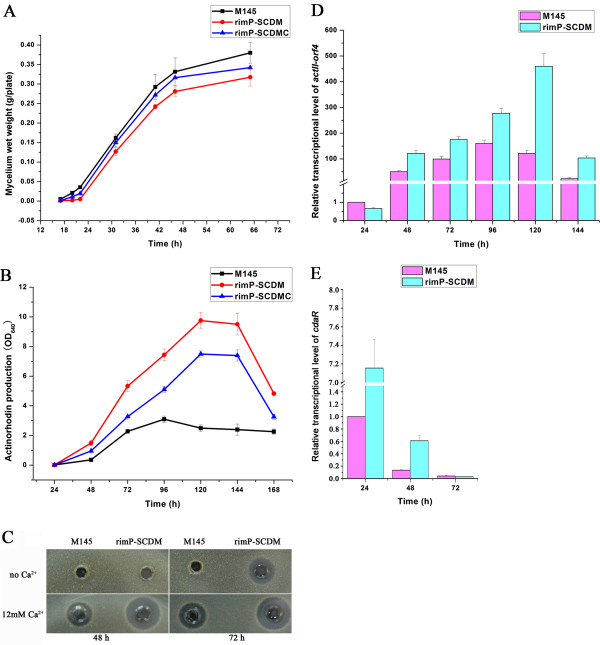
In order to clarify the function of rimP-SC in vivo, its disruption mutant (rimP-SCDM) was constructed via homologous recombination in S. coelicolor M145. Disruption of rimP-SC reduced the growth rate of S. coelicolor at initial period of rapid growth in GYM medium (Figure 2A). Interestingly, the ACT production of rimP-SCDM was remarkably increased 3-fold compared with the wild-type strain (Figure 2B). The growth rate and ACT production of the rimP-SC complemented strain (rimP-SCDMC) lay between the wild-type strain and rimP-SCDM (Figure 2A and B), suggesting that disruption of rimP-SC is the key reason for the enhancement of ACT production. Unlike ACT production, RED production of rimP-SCDM was similar to that of M145 in GYM medium (data not shown). Disruption of rimP-SC also enhanced CDA production in DNA medium (Figure 2C).
Figure 2.
Effects of rimP-SC disruption on biomass and production of actinorhodin and calcium-dependent antibiotics in S. coelicolor. (A) Growth curve of S. coelicolor M145, rimP-SCDM and rimP-SCDMC. Biomass was calculated by mycelium wet weight in GYM plate. (B) Production of actinorhodin of S. coelicolor M145, rimP-SCDM and rimP-SCDMC in GYM medium. Cell cultures (50 mg) at each time point were treated with KOH (final concentration, 1 N) and the OD640 corresponding to 10 mg of mycelium was determined. (C) Bioassay of CDA in M145 and rimP-SCDM grown in DNA medium. (D) Transcriptional analysis of actII-ORF4 by real-time RT-PCR. The transcriptional level of actII-ORF4 was detected at 24, 48, 72, 96, 120 and 144 h in M145 and rimP-SCDM grown in GYM medium. (E) Transcriptional analysis of cdaR by real-time RT-PCR. The transcriptional level of cdaR was detected at 24, 48 and 72 h in M145 and rimP-SCDM grown in DNA medium. Data were presented as the averages of the results of three independent experiments in triplicate. Error bars showed standard deviations.
To examine the function of rimP homologues in other streptomycetes, the rimP homologous gene rimP-SV was disrupted in S. venezuelae. As in S. coelicolor, disruption of rimP-SV remarkably reduced the growth rate of S. venezuelae in liquid MYM medium. And the growth rate of complemented strain (rimP-SVDMC) was almost restored to the level of the wild-type strain (Figure 3A). HPLC analysis showed that the production of jadomycin B had 2–3 folds increase in the rimP-SV disruption mutant (rimP-SVDM) (Figure 3B and C). Meanwhile, the jadomycin B production in the complemented strain was almost restored to the wild-type level, albeit slightly higher than the wild-type strain (Figure 3B and C). The disruption and complementation experiments suggested that rimP-SV was a key determinant in jadomycin B biosynthesis. Transcriptional analysis further confirmed that the increment of jadomycin production was due to the increased transcription of jadomycin biosynthetic genes (Figure 3D). Thus, the stimulatory effect on antibiotics production owing to the disruption of rimP homologues is not confined to one Streptomyces species.
Figure 3.

Effects of rimP-SV disruption on biomass and jadomycin B production. (A) Growth curve of the S. venezuelae wild-type strain (WT), rimP-SVDM and rimP-SVDMC with ethanol (ET) induction. Biomass was calculated by mycelium dry weight. (B) The yield of jadomycin B (JdB) in WT, rimP-SVDM and rimP-SVDMC. (C) HPLC analyses of fermentation filtrates from WT, rimP-SVDM and rimP-SVDMC grown for 48 h. mAU, milli absorbance units. (D) Transcriptional analysis of jadomycin B biosynthetic gene cluster by RT-PCR. Total mycelial RNA extracted after 24 h fermentation was used as template.
Transcriptional analysis of actII-ORF4, redD and cdaR
To explain the reasons for the enhanced production of several distinct antibiotics in rimP-SCDM, the transcription of corresponding biosynthetic genes were measured by real-time RT-PCR. The transcriptional levels of their pathway specific regulatory genes (actII-ORF4, redD, cdaR) involved in the biosynthesis of three well-known antibiotics (ACT, RED and CDA) were determined in M145 and rimP-SCDM. The transcription of actII-ORF4 reached the highest level in rimP-SCDM at 120 h and was 3-fold higher than that in M145 (Figure 2D). Consistent with ACT production and transcription of actII-ORF4, the transcriptional levels of SCO5072, SCO5082, SCO5086 and SCO5087 involved in ACT biosynthesis were also increased in rimP-SCDM (data not shown). Consistent with RED production, the transcriptional level of redD had no significant difference between rimP-SCDM and M145 (data not shown). The transcriptional level of cdaR in rimP-SCDM exceeded 6-fold more than that in M145 at 24 h, and the difference was narrowed from 72 h to 120 h (Figure 2E). Through the transcriptional analysis of the pathway-specific regulatory genes actII-ORF4/cdaR and biosynthetic genes involved in the ACT/CDA production, we might conclude that rimP-SC affects the ACT/CDA biosynthesis by controlling the transcription of pathway-specific regulatory gene actII-ORF4/cdaR and ACT/CDA biosynthetic genes.
RimP affects the translational efficiency and fidelity in E. coli
As a ribosome assembly cofactor, RimP may affect translational efficiency. Thus, the translational accuracy was measured using the mutated xylE as a reporter which contains a UGA stop codon instead of a UGG tryptophan codon at 47 position. When the stop codon was decoded by a near-cognate tRNA, the full length catechol dioxygenase was expressed and showed enzyme activity. To check the expression of catechol dioxygenase, all recombinant strains (BW25113/pSET152::rrnFp::xylE, rimPDM/pSET152::rrnFp::xylE, BW25113/pSET152::rrnFp::xylE* and rimPDM/pSET152::rrnFp::xylE*) were cultured in LB medium at 37°C. Under this condition, the growth rate of BW25113 was a little faster than rimPDM (Figure 4A). Meanwhile, the expression level of wild-type catechol dioxygenase in BW25113 was almost the same as rimPDM (Figure 4B). When introducing a UGA stop codon into the wild-type xylE (named as xylE*), the catechol dioxygenase activity decreased 3 orders of magnitude. Meanwhile, the activity of XylE* decreased almost 2–4 folds in rimPDM compared with BW25113 (Figure 4B). As shown in Figure 4B, the translational error rate of mutated catechol dioxygenase was about 5 × 10-4 in BW25113 and 1-2 × 10-4 in rimPDM. Therefore, the presence of RimP remarkably increased misreading of the UGA stop codon probably by decoding the near-cognate tRNA. These results implied that the presence of RimP might facilitate misreading of codons to result in the fast growth during exponential phase, but decrease the translational accuracy.
Figure 4.

Activity analysis of catechol dioxygenase in E. coli. (A) Growth curve of four different E. coli strains. (B) Activity analysis of the wild-type or mutated catechol dioxygenase. Data were presented as the averages of the results of three independent experiments in triplicate. Error bars showed standard deviations. The asterisk (*) indicates mutated catechol dioxygenase (UGG codon as tryptophan at 47 position was replaced by UGA as stop codon).
Disruption of rimP-SC significantly enhanced the expression of MetK
To study how disruption of rimP-SC enhance ACT/CDA production in S. coelicolor, transcriptions of six global activator genes (absR1, adpA, afsR, atrA, metK and rnc) and three global repressor genes (phoP, ndgR and ssgA) as well as five sigma factor genes (bldN, sigE, sigH, sigR and sigT) were analyzed by real-time RT-PCR. The results indicated that the transcriptions of five activator genes (absR1, adpA, afsR, atrA and rnc) were increased and the transcriptions of three repressor genes (phoP, ndgR and ssgA) were decreased in rimP-SCDM (Figure 5). Although the transcriptional changes of these genes could explain the increase of ACT production, it was unclear whether rimP-SC affected antibiotics biosynthesis at the translational level. Therefore, metK and sigR, whose transcription was not changed significantly in comparison with the wild-type strain, were selected for further studying their translations. To check the syntheses of MetK and SigR, the flag-tagged system was used. The result of western blotting showed that the expression of MetK was much stronger in rimP-SCDM than M145 from 24 h to 72 h (Figure 6A). However, the transcription of metK in rimP-SCDM did not exceed M145 (Figure 6B). In addition, the expression of SigR did not show significant difference between the wild-type strain and rimP-SCDM (data not show). In agreement with previous report [15], our results also showed that over-expression of MetK obviously stimulated ACT production in advance and led to an increase of ACT production up to 140% compared with the control strain M145/pIJ10500 (Figure 6C). Therefore, disruption of rimP-SC increased translational level of proteins related to secondary metabolites, such as MetK which may be one example of many proteins. The complicated mechanism that disruption of rimP homologues led to enhanced production of antibiotics is intriguing, but remains unclear, so it is worthy of studying and is being explored deeply in our lab at present.
Figure 5.
Transcriptional analysis of genes encoding global activators and repressors as well as sigma factors by real-time RT-PCR. The transcriptional levels were detected after fermentation for 24, 48, 72, 96 and 120 h in GYM medium. Data were presented as the averages of the results of three independent experiments in triplicate. Error bars showed standard deviations.
Figure 6.
Effect of rimP-SC disruption on MetK expression. (A) Detection of MetK by western blotting analysis. (B) Transcriptional analysis of metK by real-time RT-PCR. (C) The production of actinorhodin. Cell cultures (50 mg) at each time point were treated with KOH (final concentration, 1 N) and the OD640 corresponding to 10 mg of mycelium was determined. Data were presented as the averages of the results of three independent experiments in triplicate. Error bars showed standard deviations.
RimP-SCDM improved the production of polyoxin and nikkomycin
Recombinant strains containing the entire polyoxin or nikkomycin biosynthetic gene cluster were cultured for 5 days and their fermentation broths were measured by bioassay. The results showed that the fermentation broth of rimP-SCDM/pPOL had obvious stronger bioactivity than that of M145/pPOL against the indicator strain A. longipes (Figure 7A). Meanwhile, the rimP-SCDM/pPOL and M145/pPOL had comparable growth rates and final biomass (data not shown). Subsequently, we checked the transcription of polR involved in the biosynthesis of polyoxin. The results showed that the transcription of polR in rimP-SCDM/pPOL was 8-fold higher than that in M145/pPOL at 24 h (Figure 7B), which were consistent with the results of bioassay. Meanwhile, the fermentation broth of rimP-SCDM/pNIK had obvious stronger bioactivity than that of M145/pNIK against the indicator strain A. longipes (Figure 7C). The transcription of sanG encoding a positive regulator for nikkomycin biosynthesis was measured. As expected, the transcription of sanG in the rimP-SCDM/pNIK was increased 2.5-fold compared with M145/pNIK at 24 h (Figure 7D). Above results showed that rimP-SCDM could significantly improve the yield of polyoxin and nikkomycin. Therefore, it is possible that rimP-SCDM can be used as the promising heterologous expression host.
Figure 7.
Heterologous expression and transcriptional analysis of polyoxin and nikkomycin biosynthetic gene clusters. (A) Bioassay of polyoxin. (B) Transcriptional analysis of polR by real-time RT-PCR. (C) Bioassay of nikkomycin. (D) Transcriptional analysis of sanG by real-time RT-PCR. Data were presented as the averages of the results of three independent experiments in triplicate. Error bars showed standard deviations.
Discussion
SCO5703 is the homologue of RimP which facilitates the maturation of the 30S subunit in E. coli. Since RimP affects the formation of polysomes in E. coli[2], it is possible that disruption of rimP-SC also reduces the formation of polysomes and leads to the production of 30S subunit containing immature 16S rRNA in S. coelicolor. In addition, RimP-SC was involved in the antibiotics production in S. coelicolor. We postulate that the difference in the growth rate and antibiotics production correlate with the amount of polysomes which affects protein translational capacity.
Ribosomal protein S12, which is located at the interface of 30S and 50S subunits and closes to the decoding center of the ribosome, is important in maintaining translational accuracy. Contact of S12 and 16S rRNA facilitates the formation of closed conformation of 30S ribosomal subunit and hampers the entrance of near-cognate tRNA during translation [16]. The closed conformation activates EF-Tu, GTPase and ribosomes to enter the translational process [17,18]. Aside from drug resistance, many S12 mutant strains show pleiotropic effects including translational hyper-accuracy, reduced growth and impaired peptide chain elongation [19]. The K88E mutation of the S12 protein causes a high level of resistance of S. coelicolor to streptomycin and stimulates the production of ACT [20]. These phenomena might be due to the increased protein synthesis during the late growth phase and the enrichment of ribosome recycling factor RRF [21]. The phenomenon that disruption of rimP-SC increases protein synthesis at the late growth phase is similar to the K88E mutant of S12 protein [22,23]. Unlike the K88E mutant, deletion of rimP-SC may not increase the stability of 70S ribosome as the rimP mutant only hindered the maturation of 30S subunit and did not result in the change of S12 protein in E. coli[2]. In addition, accumulation of ppGpp stimulates antibiotics production in S. coelicolor[24]. The increased production of ACT in S12 mutant results from higher level of ppGpp [25]. However, the amount of ppGpp had no obvious difference between M145 and rimP-SCDM in our study (data not shown), indicating that ppGpp was not the reason for improved production of ACT in RimP-SCDM. The similar phenomenon that the hyperaccurate ribosomes exhibited slightly reduced rates of GTP hydrolysis for both cognate and near-cognate ternary complexes has been reported [26]. Therefore, the reasons for stimulating antibiotics production due to rimP-SC disruption are different from the mutation of S12 protein in S. coelicolor.
The onset of protein synthesis is determined by tRNA selection. Generally, the tRNA selection is divided into an initial selection and a later proofreading process. During initial selection, cognate aminoacyl-tRNA facilitates the stabilization of a closed 30S conformation. However, near-cognate aminoacyl-tRNA, which differs from cognate tRNA by a single, subtle mismatch in codon-anticodon base-pairing and cannot be accurately distinguished on the basis of difference in the free energy of base-pairing alone, is not disadvantage for the stabilization of a closed 30S conformation [17]. Stabilization of the closed form of 30S ribosomal subunit could reduce the translational fidelity and increase the translational speed. The translational accuracy was measured using xylE gene as a reporter in E. coli wild-type strain and rimPDM. The results showed that RimP might stabilize the closed form and accelerate the reconstitution of 30S ribosomal subunit induced by cognate or near-cognate tRNA, thus speeding up the translation. Without RimP, the closed 30S form might be unstable and unfavorable for selection of near-cognate tRNA, thus leading to higher translation fidelity and lower translation speed.
Conclusions
As in E. coli, RimP-SC encoded a cofactor involved in ribosome assembly of the 30S subunit and its disruption reduced growth rate at initial period of rapid growth in S. coelicolor. RimP-SC also played an essential role in actinorhodin and calcium-dependent antibiotics production. Disruption of rimP-SC enhanced expression of MetK and protein translational accuracy, resulting in increased antibiotics production. This is the first study to address the relationship between ribosome assembly cofactor and antibiotics production. Our results provided an approach for yield improvement based on rimP homologues disruption, which was also effective in S. venezuelae, implying that the approach might be adopted to increase antibiotics production in other Streptomyces species. Ultimately, more peptidyl nucleoside antibiotics—polyoxin and nikkomycin could be produced in rimP-SC disruption mutant than M145, indicating that rimP-SC disruption mutant could be used as a promising host for heterologous expression.
Materials and methods
Bacterial strains, plasmids, primers, growth conditions and assay of antibiotics
Bacterial strains and plasmids used in this study were listed in Table 1. The primers used in this study were listed in Table 2. E. coli JM109 was used as a general host for routine cloning experiment. BW25113 was used as an E. coli host for the construction of rimP disruption mutant via λ-Red-mediated recombination technology. E. coli ET12567/pUZ8002 was used as a host for transferring DNA from E. coli to Streptomyces by intergeneric conjugation. S. coelicolor M145 was a derivative of the wild-type strain S. coelicolor A3(2) lacking plasmids SCP1 and SCP2. S. venezuelae ATCC10712 was the wild-type strain of S. venezuelae. Alternaria longipes was the indicator strain of polyoxin and nikkomycin. Staphylococcus aureus was the indicator strain of calcium-dependent antibiotics (CDA). Usually, E. coli and its derivatives were grown at 37°C. Streptomyces and their derivatives were grown at 28°C. Yeast extract-malt extract liquid medium (YEME), GYM medium, Difco Nutrient Agar (DNA) medium supplemented with 0.5% NaCl (w/v) and agar minimal medium (MM) supplemented with mannitol as sole carbon source were prepared for growth and sporulation of Streptomyces as described previously [6]. SP medium (3% mannitol, 1% soluble starch, 0.75% yeast extract, and 0.5% soy peptone, pH 6.0) for polyoxin and nikkomycin production was prepared as described previously [27]. Potato dextrose agar medium (PDA) for the detection of polyoxin and nikkomycin was used. MYM medium for jadomycin production was prepared as described previously [28]. For the production of ACT and RED, fresh spores of S. coelicolor were inoculated in GYM medium as described previously [6]. ACT and RED were detected spectrophotometrically [6]. CDA production in DNA medium was assayed as described previously [29]. Jadomycin production was measured as described previously [28,30,31]. For fermentation, spore suspensions of S. venezuelae were inoculated in MYM and grown at 28°C on a rotary shaker (220 rpm) for 24 h as seed cultures. 5 ml (5% v/v) of seed culture was transferred into flasks containing 100 ml of galactose-isoleucine medium with 1.5 ml ethanol, and then cultured at 28°C on a rotary shaker (220 rpm) for jadomycin production. Jadomycin B was identified by HPLC analysis (Agilent 1260 HPLC and RPC-18) at 316 nm absorption wavelengths. Chemical reagent, mobile phase and gradient elution process were performed as described previously [31].
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strains/plasmids | Relevant characteristics | Source/references |
|---|---|---|
|
Streptomyces |
|
|
|
S. coelicolor M145 |
A derivative of the wild-type strain S. coelicolor A3(2) lacking plasmids SCP1 and SCP2 |
[6] |
| rimP-SCDM |
The rimP-SC disruption mutant of S. coelicolor |
This study |
| rimP-SCDMC |
The complemented strain of rimP-SCDM |
This study |
| M145/pNIK |
S. coelicolor M145 containing the entire nikkomycin biosynthetic gene cluster |
This study |
| rimP-SCDM/pNIK |
rimP-SCDM containing the entire nikkomycin biosynthetic gene cluster |
This study |
| M145/pPOL |
S. coelicolor M145 containing the entire polyoxin biosynthetic gene cluster |
This study |
| rimP-SCDM/pPOL |
rimP-SCDM containing the entire polyoxin biosynthetic gene cluster |
This study |
|
S. venezuelae ATCC10712 |
The wild-type strain of S. venezuelae |
[6] |
| rimP-SVDM |
The rimP-SV disruption mutant of S. venezuelae |
This study |
| rimP-SVDMC |
The complemented strain of rimP-SVDM |
This study |
|
E. coli |
|
|
| JM109 |
recA1, endA1, gyrA96, thi-1, hsdR17, supE44, relA1, Δ(lac-proAB)/F’[traD36, proAB+, lacIq, lacZΔM15] |
Invitrogen |
| BW25113 |
K-12 derivative; ΔaraBAD ΔrhaBAD |
[32] |
| ET12567/pUZ8002 |
dam dcm hsdS cat tet/pUZ8002 |
[33] |
| rimPDM |
The rimP disruption mutant of E. coli BW25113 |
This study |
| rimPDMC |
The heterologous complemented strain of rimPDM |
This study |
| rimPDM/pSET152::rrnFp::SCO5703 |
rimPDM containing pSET152::rrnFp::SCO5703 for heterologous complementation analysis |
This study |
| BW25113/pSET152::rrnFp::xylE |
BW25113 containing pSET152::rrnFp::xylE for catechol dioxygenase assay |
This study |
| BW25113/pSET152::rrnFp::xylE* |
BW25113 containing pSET152::rrnFp::xylE* for catechol dioxygenase assay |
This study |
| rimPDM/pSET152::rrnFp::xylE |
rimPDM containing pSET152::rrnFp::xylE for catechol dioxygenase assay |
This study |
| rimPDM/pSET152::rrnFp::xylE* |
rimPDM containing pSET152::rrnFp::xylE* for catechol dioxygenase assay |
This study |
| Plasmids |
|
|
| pRIMPSC3 |
Plasmid used for the construction of rimP-SCDM |
This study |
| pRIMPSC4 |
pSET152 containing the intact rimP-SC with its putative promoter |
This study |
| pRIMPSV3 |
Plasmid used for the construction of rimP-SVDM |
This study |
| pRIMPSV4 |
pSET152 containing the intact rimP-SV with its putative promoter |
This study |
| pSET152::rrnFp::xylE |
pSET152 containing the wild-type xylE and the promoter of rrnF for activity detection of catechol dioxygenase |
This study |
| pSET152::rrnFp::xylE* |
pSET152 containing the mutated xylE and the promoter of rrnF for activity detection of catechol dioxygenase |
This study |
| pIJ10500::metK |
pIJ10500 containing the intact metK with its promoter |
This study |
| pIJ10500::sigR |
pIJ10500 containing the intact sigR with its promoter |
This study |
| pNIK |
pSET152 containing the entire nikkomycin biosynthetic gene cluster |
[34] |
| pPOL | pSET152 containing the entire polyoxin biosynthetic gene cluster | [35] |
Table 2.
Primers used in this study
| Genes and primers | Sequence (5′-3′) |
|---|---|
|
rimP homologues relevant primers |
|
| Lrimp-SC-F |
CCCAAGCTTGGCCAGCCGGTCCTCCAGTT |
| Lrimp-SC-R |
GCTCTAGAGGTGGTGCTCATCCGGGTGA |
| Rrimp-SC-F |
GGGGTACCAGGCCCACCACCCGCAGACT |
| Rrimp-SC-R |
GGAATTCCGGCGTGCGGCTTGGATCTA |
| CrimP-SC-F |
CGCGGCCAGTTCCTCACTGT |
| CrimP-SC-R |
CAGGGCGCTCATGTCGATGT |
| ECrimP-F |
TTGTCCACATTAGAGCAAAAATTAACAGAGATGATTACTGGAATTGTGAGCGGATAAC |
| ECrimP-R |
TCTGGATATTACTCAGCGCGAACACTTCATCTTTACCTTAGGCGATTAAGTTGGGTAA |
| YZECrimP-F |
TTGTCCACATTAGAGCAAA |
| YZECrimP-R |
TTAAAAGTGGGGAACCAG |
| PFrimP-F |
CGGGATCCGACCCACAACAGCACACG |
| PFrimP-R |
GCTCTAGATCTCCTTCTCCCGTACCAA |
| LrimP-SV-F |
AAGCTTGCCGAACGGTACAGAAAGGGTA |
| LrimP-SV-R |
TCTAGATCGCTCTGGGTGGTGCTCATC |
| RrimP-SV-F |
GGATCCGGCGAGTACGTCCTCGAAGT |
| RrimP-SV-R |
GATATCACCTTGCTCTCCACACCGAACTCC |
| CrimP-SV-F |
CGGCGGTTCGAAACCCATGC |
| CrimP-SV-R |
CTACGCCTCCTCTTCCTTCTTGTCCTT |
| YZrimP-SV-F |
GTGGTACGTCGTCGAAGATC |
| YZrimP-SV-R |
GCAAAGCGTCAGTCAACTTG |
| Primers for real-time PCR of genes in S. coelicolor |
|
| RTact-F |
GCTCCTCAGGCGGCACGA |
| RTact-R |
GCCGGCGGGTGTGGTACA |
| RTred-F |
GCCCTGACGCGCTATTGG |
| RTred-R |
GGTGGTGGGCGAGACGGA |
| RTcda-F |
GGAAAGCGACGCCTACTT |
| RTcda-R |
AGGCTCGTCTTTCCGATT |
| RTmetK-F |
CGAGCCCGTGGGTCTGTT |
| RTmetK-R |
CAGGTCGAGAGCGCGGAT |
| RTsigR-F |
CGACCACCTGCCCGACTC |
| RTsigR-R |
CCCCATGATGTCCGCGAT |
| RTsigE-F |
GGAGGAGGTGCCGACCGA |
| RTsigE-R |
TTCCCGCCGACATTCCGA |
| RTsigH-F |
GGAGCCGCTGGACGACCT |
| RTsigH-R |
CACCGCCCAGCCCTTGTC |
| RTbldN-F |
GACAGCGCCCGCATGATG |
| RTbldN-R |
GAGCGCCCGCAGAAAGGT |
| RTsigT-F |
GCCCTCGTCTCCGCCTAC |
| RTsigT-R |
CAGGCGTTCGGTGTCGTC |
| RTabsR1-F |
CCCGCAGTCGATCATGGA |
| RTabsR1-R |
GCAGGGCGAACTCCTTGTC |
| RTadpA-F |
AGCACCTCCACGAGCAGTTC |
| RTadpA-R |
CGTCCACCGAGTAGTCCGA |
| RTafsR-F |
GGCGGTGGATCTGCTGTG |
| RTafsR-R |
ACATCGCTGAGAACGGTGC |
| RTatrA-F |
CCGGCGGTGCGATGAGTA |
| RTatrA-R |
ACCCCAGCTCGCCGAACA |
| RTndgR-F |
CGACGTGACGGGCGAGAG |
| RTndgR-R |
GGAGCCGGCCTTCATGGT |
| RTphoP-F |
ACGTTCCCGTGATCATGGTG |
| RTphoP-R |
CAGTACGGCTCGGATGCG |
| RTrnc-F |
GGTGATCGGCGCGGTCTA |
| RTrnc-R |
CCTTCGGTCGCGGTGAGT |
| RTssgA-F |
CAGGCGCTGTTCCGTTCC |
| RTssgA-R |
GATGCGGTCCAGGGCCTC |
| RT5072-F |
GACGACCTGCCGCTCAAG |
| RT5072-R |
GAACGATGTGCGGTGGGT |
| RT5082-F |
GGAGGCCCTGGAGCAGTC |
| RT5082-R |
GCCGGCGATGATGATCTC |
| RT5086-F |
ACCTCACCGGCGTGTTCC |
| RT5086-R |
CGTGCTTCGAGGCGGAGT |
| RT5087-F |
GAACGACCGCCACGAGAC |
| RT5087-R |
GATCTCCAGCGAGCCGAT |
| RTsanG-F |
GGCCACCCTGCAGACGTAC |
| RTsanG-R |
CGGGACAGGTCGAACGTG |
| RTpolR-F |
GGTCTCCCGCGGACAACA |
| RTpolR-R |
GCGGCTCGTAGGACGTGA |
| RThrdB-SC-F |
GATCGCCGAGTCCGTCTC |
| RThrdB-SC-R |
CACTGAGTGGCCGGAATC |
| Primers for real-time PCR of genes in S. venezuelae |
|
| RTjadR3-F |
CACGTGGACGTGACGGATACGG |
| RTjadR3-R |
GGGTGTCGGCGAGGTTTCCTTC |
| RTjadW1-F |
TCGTCTGCTCCGACATCACCC |
| RTjadW1-R |
GCAGGAAGGAGACGCTCAGGTC |
| RTjadW2-F |
ACGTACTGATCCACTGCGCCTCC |
| RTjadW2-R |
CGATCAGGGAGTGCAGCGAGG |
| RTjadW3-F |
ACTACGGCAGCAACGAGAAGGC |
| RTjadW3-R |
AGGGCGAGGGTCATCGTGTC |
| RTjadR2-F |
TCGGCGATCAGTTCGGGAGC |
| RTjadR2-R |
AGCCATTCGCCGTTGTCCC |
| RTjadR1-F |
TGGACGGCTTGGAGGTCTGC |
| RTjadR1-R |
GGCTGCTCACATGGGTGTCG |
| RTjadJ-F |
CTGTCGGAGGCTCAGAACGC |
| RTjadJ-R |
ACGATCACGTTCGCAAGCAG |
| RTjadI-F |
TGCACAGCACTCTGATCGTGG |
| RTjadI-R |
GCGTTCGCCTCCCAGTTGTAG |
| RTjadA-F |
CCCCAACACCGTGGTCTCC |
| RTjadA-R |
GCGGTCGTTCTGCTTGGTG |
| RTjadB-F |
GGAGCCAGGGCAGCCAGTAC |
| RTjadB-R |
CGAAGGTGGAGCCGTATCCG |
| RTjadC-F |
GCAGCAAGACCTTCACCCTCG |
| RTjadC-R |
CCGACAGGTGCGCGTTGAC |
| RTjadE-F |
GCCGACGAGCTGTGGAACG |
| RTjadE-R |
GAGGCCAGGTAGCCGACGAG |
| RTjadD-F |
GCCTTGCTGCACGACTACCG |
| RTjadD-R |
ACGCCGTCCTCGTTCTCCTC |
| RTjadF-F |
ACGCCGCTCTGGGTGAACT |
| RTjadF-R |
GATGTCGAGTCCTGAGACCTTGC |
| RTjadG-F |
ACCTGACCGTCTTCAACCTCTTCG |
| RTjadG-R |
TGCTGCTGGTCCGGCTTCAC |
| RTjadH-F |
GACGACGACGCCGTGGAGA |
| RTjadH-R |
GATGTCCTCGCCCGTGATGC |
| RTjadK-F |
CGGCTGCGGACAGGAGTACG |
| RTjadK-R |
GAGGCCCAGGCTGATGTTGTG |
| RTjadL-F |
GGAAGGAGGAACGGAAGGACG |
| RTjadL-R |
ATCAGGGTGTAGAGGGCGAGG |
| RTjadM-F |
CCCGCTACACCGGAGTCCC |
| RTjadM-R |
GAGTCCCGTGCCGAGTCCC |
| RTjadN-F |
GCAGGGTTTCGGTCTGGAGG |
| RTjadN-R |
CGAGGCCGTTCTGGGTGATC |
| RTjadX-F |
CCACCACCGACCTCACCG |
| RTjadX-R |
CGAAGTGGGCGGAGGGC |
| RTjadO-F |
TTCCACAAGTCCAACCGCAAC |
| RTjadO-R |
TTCGATCAGCGGCTGGGTC |
| RTjadP-F |
AAGCACGTCCTGGCCGAGAAG |
| RTjadP-R |
GGTCCATGCCGAAGGCGATGT |
| RTjadQ-F |
CGACAAGCCGATGATCTACTACCC |
| RTjadQ-R |
GCGTGAGGTTCTTGGCGATGT |
| RTjadS-F |
GTCTTCCCGCCCAACCACG |
| RTjadS-R |
GCGAGGGAGCCAGCGTCAC |
| RTjadT-F |
CGACGAGGTGTACGGCACG |
| RTjadT-R |
CAGGATGCGTTCGGTCAGG |
| RTjadU-F |
CAGGTCAATCAGGTCAGCCACA |
| RTjadU-R |
CCGGTCGGACAGGATCAGC |
| RTjadV-F |
GACGAGCCGCAGGGCGAG |
| RTjadV-R |
CCGCTCCGCCACCATCCG |
| RThrdB-SV-F |
AGATTCCGCCAACCCAGTG |
| RThrdB-SV-R |
GAGCGTCGTCTCGTCTCGTC |
| other primers |
|
| rrnFp-F |
TGGAGGGAGATACGAGAACG |
| rrnFp-R |
CCCAGAGTGAAGGGCAGATT |
| MxylE-F |
TGAACCGAAGTGGATAAGTT |
| MxylE-R |
AGCCTTCAGATAGACACGGC |
| metK-F |
CGGCGGCTGGAATGAATGACCC |
| metK-R |
CAGGCCCGCGGCCTCGCGCA |
| sigR-F |
GGGCGGAGATCAGCCAGGAAAG |
| sigR-R |
TGACCCCGAGCCTTTCGCTTCGT |
| polB-F |
GGTGAAGACGCCAACGAC |
| polB-R |
GATCGGAGCGCGTACCAG |
| sanG-F |
GGGGTACCGTGCGTCAACCTCATCCCG |
| sanG-R |
GGAATTCGCTTGCCCGCTGGTCT |
| Kan-F |
TCTAGAGATCCCCTGGATACCGCTCG |
| Kan-R | GGATCCGTACCCGAACCCCAGAGTC |
Plasmids pBluescript KS+, pEASY-Blunt and pUC119::kan were used for routine cloning experiments in E. coli. The Streptomyces—E. coli shuttle plasmid pKC1132 was used to construct gene disruption mutants via homologous recombination. The integrative plasmid pSET152 was used to introduce a single copy of rimP homologue gene into the Streptomyces chromosome. pIJ10500 containing hygromycin B (hyg) resistance gene and 3 × FLAG tag was used in western blotting experiment. The xylE from pIJ4083 was used for the construction of the reporter system. When necessary, antibiotics were used at the following concentrations: ampicillin (100 μg · ml−1), kanamycin (100 μg · ml−1), apramycin (100 μg · ml−1) or hygromycin B (50 μg · ml−1) in LB for E. coli; Nalidixic acid (25 μg · ml−1), apramycin (100 μg · ml−1) or kanamycin (100 μg · ml−1) in MS for Streptomyces[6].
Construction of the recombinant strains
The rimP disruption mutant (rimPDM) of E. coli BW25113 was constructed by PCR targeting as follows: A 1.2 kb DNA fragment containing the kanamycin resistance gene (kan) was amplified by PCR using primers ECrimP-F and ECrimP-R. This fragment covered the 38-bp upstream region and the 65-bp downstream region of rimP. Then the fragment was purified and introduced into the BW25113 by electroporation. Finally, the kanamycin resistance gene substituted the most of rimP coding region by homologous recombination. The resulting strain was confirmed by PCR amplification using primers YZECrimP-F and YZECrimP-R. In order to clarify the relationship between rimP of E. coli and SCO5703 of S. coelicolor, the heterologous complemented strain was constructed according to the following steps: Firstly, promoter region of rrnF was amplified with primer rrnFp-F and rrnFp-R from S. coelicolor and ligated into pEASY-blunt to generate pEASY-blunt-rrnFp. The authenticity of PCR amplicon was verified by sequencing, and then it was ligated into the NotI-BamHI site of integrative vector pSET152 to give pSET152::rrnFp. Meanwhile, the DNA fragment containing the intact SCO5703 was amplified by PCR using primers PFrimP-F and PFrimP-R, then it was digested with XbaI-BamHI and ligated into the corresponding sites of pSET152::rrnFp to generate pSET152::rrnFp::SCO5703. Finally, rimPDM was transformed with the plasmid pSET152::rrnFp::SCO5703 to generate the heterologous complemented strain of rimPDM (rimPDMC).
To construct the rimP-SC disruption mutant (rimP-SCDM) of S. coelicolor M145, the DNA fragment corresponding to the upstream region of rimP-SC (extending from positions −1269 to +12 with respect to the rimP-SC translation start codon) was amplified by PCR using primers LrimP-SC-F and LrimP-SC-R and inserted into the HindIII-XbaI sites of pUC119::kan to generate pRIMPSC1. The DNA fragment corresponding to the downstream region of rimP-SC (extending from positions +403 to +1576 with respect to the rimP-SC translation start codon) was amplified by PCR using primers RrimP-SC-F and RrimP-SC-R and inserted into the KpnI-EcoRI sites of pRIMPSC1. The resulting plasmid pRIMPSC2 was then digested with HindIII and EcoRI. A 3.3 kb DNA fragment was isolated and ligated into the corresponding sites of pKC1132 to give pRIMPSC3. The authenticity of all PCR amplicons was verified by sequencing. Subsequently, pRIMPSC3 was introduced into S. coelicolor M145 via ET12567/pUZ8002 by conjugal transfer and the transformants conferring kanamycin resistance (Kanr) and apramycin sensitivity (Aprs) were selected, and they were further confirmed by PCR using primers LrimP-SC-F and RrimP-SC-R. For complementation analysis, the fragment containing the intact rimP-SC with its putative promoter region was amplified with primers CrimP-SC-F and CrimP-SC-R and inserted into the EcoRV site of pSET152 to generate pRIMPSC4. Subsequently, pRIMPSC4 was introduced into rimP-SCDM by conjugal transfer and the complemented strain was confirmed by PCR.
To construct the rimP-SV disruption mutant (rimP-SVDM) of S. venezuelae ATCC10712, the DNA fragment corresponding to the upstream region of rimP-SV was amplified by PCR using primers LrimP-SV-F and LrimP-SV-R and inserted into the HindIII-XbaI sites of pKC1139 to generate pRIMPSV1. The DNA fragment corresponding to the downstream region of rimP-SV was amplified by PCR using primers RrimP-SV-F and RrimP-SV-R and inserted into the BamHI-EcoRV sites of pRIMPSV1 to generate pRIMPSV2. Kanamycin resistance gene was amplified by PCR using primers Kan-F and Kan-R and inserted into the BamHI-XbaI sites of pRIMPSV2 to generate pRIMPSV3. The authenticity of all PCR amplicons was verified by sequencing. Subsequently, pRIMPSV3 was introduced into S. venezuelae ATCC10712 via ET12567/pUZ8002 by conjugal transfer and transformants conferring kanamycin resistance (Kanr) and apramycin sensitivity (Aprs) were selected, and they were further confirmed by PCR using primers YZrimP-SV-F and YZrimP-SV-R. For complementation analysis, the fragment containing the intact rimP-SV with its putative promoter region was amplified using primers CrimP-SV-F and CrimP-SV-R and inserted into the EcoRV site of pSET152 to generate pRIMPSV4. Subsequently, pRIMPSV4 was introduced into rimPSV-DM by conjugal transfer and the complemented strain was confirmed by PCR.
For detection of MetK and SigR expression in S. coelicolor, the 3 × FLAG-tagged system was applied and series of plasmids were constructed as follows: The DNA fragment containing the intact metK or sigR with its respective promoter was amplified by PCR with primers metK-F/metK-R or sigR-F/sigR-R and ligated into the StuI site of pIJ10500 to generate pIJ10500::metK or pIJ10500::sigR. The resulting plasmid pIJ10500::metK or pIJ10500::sigR was introduced into S. coelicolor M145 and rimP-SCDM by conjugal transfer, respectively. All the recombinant strains were subsequently confirmed by PCR amplification.
RNA isolation, RT-PCR and real-time RT-PCR
Total RNA were isolated from Streptomyces as described previously [36,37]. For reverse transcription PCR (RT-PCR) and quantitative real-time reverse transcription PCR (real-time RT-PCR), the genomic DNA was removed from RNA samples with RQ1 RNase-free DNase (Promega), the synthesis of the first-strand cDNA was performed with Superscript III first-strand Synthesis System (Invitrogen) as described previously [38]. Reaction mixtures contained 6 pmol of random primers (Invitrogen) and 1 μg of RNA in a total volume of 20 μl. The reverse transcription conditions were as follows: 65°C for 5 min, 25°C for 5 min, 50°C for 45 min, 55°C for 45 min, and 72°C for 10 min. RT-PCR reaction parameters were as follows: 95°C for 5 min, followed by 30 amplification cycles consisting of 95°C for 30 seconds denaturation, 55°C for 30 seconds annealing, 72°C for 45 seconds extension and a final extension of 72°C for 10 min. RT-PCR was performed without reverse transcriptase to test for DNA contamination in the RNA samples. After 30 cycles of amplification, the products were displayed on a 2% agarose gel and visualized by staining with ethidium bromide. Real-time RT-PCR was performed in 96-well rotor using the Eppendorf Realplex system, and the reaction mixtures were prepared as follows: Each reaction (50 μl) contained 0.1-10 ng of cDNA, 25 μl Power SYBR Green PCR master mix (Toyobo, QPS-201), and 0.4 μM of forward and reverse primers respectively. The reaction conditions were maintained at 95°C for 30 seconds, followed by 40 amplification cycles consisting of 15 seconds denaturation at 95°C, 20 seconds annealing at 60°C and 30 seconds extension at 72°C. Fluorescence was measured at the end of each cycle. The final dissociation stage was run to generate a melting curve and consequently verify the specificity of the amplification products. Changes in levels of gene expression were calculated automatically with the Detection Software using the ΔΔCT method. The hrdB was used as the housekeeping gene reference for RT-PCR and real-time RT-PCR.
Construction of the xylE reporter system and Detection of translational fidelity
The xylE was isolated from pIJ4083 by BglII and BamHI digestions, and then it was inserted into the BamHI site of pSET152 to generate pSET152::xylE. The DNA fragment containing the rrnF promoter from pEASY-blunt-rrnFp was isolated and inserted in the upstream of xylE in pSET152 to generate pSET152::rrnFp::xylE. For the construction of mutated xylE reporter plasmid, pSET152::rrnFp::xylE was used as the template for PCR amplification with primers MxylE-F and MxylE-R. The authenticity of PCR amplicon was verified by sequencing. The mutated plasmid pSET152::rrnFp::xylE*, which contained alterations in the 5′ region of the xylE gene, was introduced a premature stop codon that abolished catechol dioxygenase activity. The reporter plasmids pSET152::rrnFp::xylE and pSET152::rrnFp::xylE* had the correct orientation of promoter in favor of transcriptional detection of xylE, and then both of them were introduced into BW25113 and rimPDM to estimate translational fidelity. The translational error rate was calculated as activity of the wild-type catechol dioxygenase divided by that of mutated catechol dioxygenase in the same strain.
Activity assays of XylE
The detailed steps for activity detection of catechol dioxygenase were performed as described previously with minor revised [6]. For the recovery of recombinant strains, they were inoculated in 3 ml of LB and incubated for 8–10 h at 37°C with shaking at 220 rpm. Then, the same amount of cells of each strain was transferred to 50 ml of LB medium and incubated for 3.5, 5.5, 7, 9, 12 or 23 h, respectively. Cultures of 1 ml were harvested. After washing with 1 ml sample buffer (100 mM phosphate buffer pH 7.5, 20 mM EDTA pH 8.0, 10% acetone), they were re-suspended in 0.5 ml of sample buffer. The 0.5 ml of cell suspension was sonicated on ice and 5 μl of 10% Triton X-100 was added. It was placed on ice for 15 min and centrifuged for 10 min at 12,000 rpm, the supernatants were used for activity assays of XylE. The reaction mixture for measurement of catechol dioxygenase activity consisted of 0.5 ml of assay buffer (10 mM phosphate buffer, pH 7.5, 0.2 mM catechol) and 5–50 μl of cell extract. Protein concentrations of cell extracts were measured according to the BCA protein assay method by using BSA as the standard. The catechol dioxygenase activity was calculated as the rate of change in optical density at 375 nm per minute per milligram of protein. The formula is as follows: catechol dioxygenase (mU) = 30.03 × △A375/time (min).
Western blotting of MetK and SigR in S. coelicolor
For western blotting analysis, cell extracts from M145/pIJ10500::metK, M145/pIJ10500::sigR, rimP-SCDM/pIJ10500::metK and rimP-SCDM/pIJ10500::sigR, grown in the GYM medium at different time points, were sonicated on ice. The concentration of total protein was determined by BCA protein assay using BSA as the standard sample. Equal concentrations of proteins (50 μg) from different time-point samples were loaded onto 12% polyacrylamide/SDS gel electrophoresis. Proteins in the gels were transferred to PVDF western blotting membranes (Roche, Germany) and probed with monoclonal ANTI-FLAG M2 antibody (Sigma-aldrich, USA) as recommended by the manufacturer. The antibodies on the membranes were hybridized with the goat Anti-Mouse IgG-HRP as secondary antibody (Jackson, USA) and the position of tagged-FLAG was visualized through cECL western blot kit (CWBIO Corporation, China).
Heterologous expression and bioassays of polyoxin and nikkomycin
The entire polyoxin and nikkomycin biosynthetic gene clusters were ligated with integrated vector pSET152 to generate pPOL and pNIK, respectively [34,35]. Then they were introduced into M145 and rimP-SCDM to generate recombinant strains M145/pPOL, rimP-SCDM/pPOL, M145/pNIK and rimP-SCDM/pNIK. Recombinant strains containing the entire polyoxin or nikkomycin biosynthetic gene cluster were confirmed by PCR amplification using primers polB-F/polB-R or sanG-F/sanG-R respectively. Then 5 days’ fermentation broths of all the recombinant strains were measured by a disk agar diffusion method using A. longipes as indicator strain.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YP and CL performed the experiments of S. coelicolor and S. venezuelae respectively. HD carried out the HPLC analysis of ppGpp. LY assisted with experiments. YP wrote the draft manuscript. GL and HT supervised the whole work and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yuanyuan Pan, Email: panyy@im.ac.cn.
Cheng Lu, Email: lucheng107@163.com.
Hailing Dong, Email: donghl@im.ac.cn.
Lingjun Yu, Email: Yulj_113@yahoo.com.cn.
Gang Liu, Email: liug@im.ac.cn.
Huarong Tan, Email: tanhr@im.ac.cn.
Acknowledgements
We thank Professor Mervyn Bibb (John Innes Centre, Norwich, UK) for critical reading in preparation of this manuscript and Chris D. Den Hengst for providing pIJ10500 (John Innes Centre, Norwich, UK). This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31030003, 31200929 and 31270110) and Ministry of Science and Technology of China (Grant No. 2009CB118905).
References
- Neidhardt F, Ingraham J, Schaechter M. Physiology of the Bacterial Cell. A Molecular Approach. Sunderland, MA: Sinauer Associates, Inc, Publishers; 1990. [Google Scholar]
- Nord S, Bylund GO, Lovgren JM, Wikstrom PM. The RimP protein is important for maturation of the 30S ribosomal subunit. J Mol Biol. 2009;386:742–753. doi: 10.1016/j.jmb.2008.12.076. [DOI] [PubMed] [Google Scholar]
- Bunner AE, Nord S, Wikstrom PM, Williamson JR. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J Mol Biol. 2010;398:1–7. doi: 10.1016/j.jmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology. 1999;145(Pt 9):2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, England: John Innes Foundation; 2000. [Google Scholar]
- Malpartida F, Hopwood DA. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature. 1984;309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- Cerdeno AM, Bibb MJ, Challis GL. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem Biol. 2001;8:817–829. doi: 10.1016/S1074-5521(01)00054-0. [DOI] [PubMed] [Google Scholar]
- Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BA, Hayes MA. et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol. 2002;9:1175–1187. doi: 10.1016/S1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- Corre C, Challis GL. Evidence for the unusual condensation of a diketide with a pentulose in the methylenomycin biosynthetic pathway of Streptomyces coelicolor A3(2) ChemBioChem. 2005;6:2166–2170. doi: 10.1002/cbic.200500243. [DOI] [PubMed] [Google Scholar]
- Challis GL. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology. 2008;154:1555–1569. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto-Hosoya Y, Sato TA, Ochi K. Resistance to paromomycin is conferred by rpsL mutations, accompanied by an enhanced antibiotic production in Streptomyces coelicolor A3(2) J Antibiot. 2000;53:1424–1427. doi: 10.7164/antibiotics.53.1424. [DOI] [PubMed] [Google Scholar]
- Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K. Ribosome engineering and secondary metabolite production. Adv Appl Microbiol. 2004;56:155–184. doi: 10.1016/S0065-2164(04)56005-7. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Lezhava A, Hosaka T, Okamoto-Hosoya Y, Ochi K. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3(2) J Bacteriol. 2003;185:601–609. doi: 10.1128/JB.185.2.601-609.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol. 2007;374:1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/S0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Wang G, Inaoka T, Okamoto S, Ochi K. A novel insertion mutation in Streptomyces coelicolor ribosomal S12 protein results in paromomycin resistance and antibiotic overproduction. Antimicrob Agents Chemother. 2009;53:1019–1026. doi: 10.1128/AAC.00388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh A, Ochi K. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J Antibiot. 1997;50:532–535. doi: 10.7164/antibiotics.50.532. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Xu J, Ochi K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol Microbiol. 2006;61:883–897. doi: 10.1111/j.1365-2958.2006.05285.x. [DOI] [PubMed] [Google Scholar]
- Okamoto-Hosoya Y, Hosaka T, Ochi K. An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2) Microbiology. 2003;149:3299–3309. doi: 10.1099/mic.0.26490-0. [DOI] [PubMed] [Google Scholar]
- Tamehiro N, Hosaka T, Xu J, Hu H, Otake N, Ochi K. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl Environ Microbiol. 2003;69:6412–6417. doi: 10.1128/AEM.69.11.6412-6417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Tanaka Y, Ochi K. The G243D mutation (afsB mutation) in the principal sigma factor sigmaHrdB alters intracellular ppGpp level and antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 2010;156:2384–2392. doi: 10.1099/mic.0.039834-0. [DOI] [PubMed] [Google Scholar]
- Wang G, Hosaka T, Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol. 2008;74:2834–2840. doi: 10.1128/AEM.02800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Rodnina MV. Optimization of speed and accuracy of decoding in translation. EMBO J. 2010;29:3701–3709. doi: 10.1038/emboj.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Tan H, Li J. Cloning and function of sanQ: a gene involved in nikkomycin biosynthesis of Streptomyces ansochromogenes. Curr Microbiol. 2002;45:175–179. doi: 10.1007/s00284-001-0115-4. [DOI] [PubMed] [Google Scholar]
- Yang K, Han L, He J, Wang L, Vining LC. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene. 2001;279:165–173. doi: 10.1016/S0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- Anderson TB, Brian P, Champness WC. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol Microbiol. 2001;39:553–566. doi: 10.1046/j.1365-2958.2001.02240.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci U S A. 2009;106:8617–8622. doi: 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doull JL, Singh AK, Hoare M, Ayer SW. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J Ind Microbiol. 1994;13:120–125. doi: 10.1007/BF01584109. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Li J, Li L, Yang H, Tian Y, Tan H. Cloning, reassembling and integration of the entire nikkomycin biosynthetic gene cluster into Streptomyces ansochromogenes lead to an improved nikkomycin production. Microb Cell Fact. 2010;9:6. doi: 10.1186/1475-2859-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li L, Feng C, Chen Y, Tan H. Novel polyoxins generated by heterologously expressing polyoxin biosynthetic gene cluster in the sanN inactivated mutant of Streptomyces ansochromogenes. Microb Cell Fact. 2012;11:135. doi: 10.1186/1475-2859-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Tian Y, Yang H, Tan H. A pathway-specific transcriptional regulatory gene for nikkomycin biosynthesis in Streptomyces ansochromogenes that also influences colony development. Mol Microbiol. 2005;55:1855–1866. doi: 10.1111/j.1365-2958.2005.04512.x. [DOI] [PubMed] [Google Scholar]
- Pan Y, Liu G, Yang H, Tian Y, Tan H. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol Microbiol. 2009;72:710–723. doi: 10.1111/j.1365-2958.2009.06681.x. [DOI] [PubMed] [Google Scholar]
- Pan Y, Wang L, He X, Tian Y, Liu G, Tan H. SabR enhances nikkomycin production via regulating the transcriptional level of sanG, a pathway-specific regulatory gene in Streptomyces ansochromogenes. BMC Microbiol. 2011;11:164. doi: 10.1186/1471-2180-11-164. [DOI] [PMC free article] [PubMed] [Google Scholar]