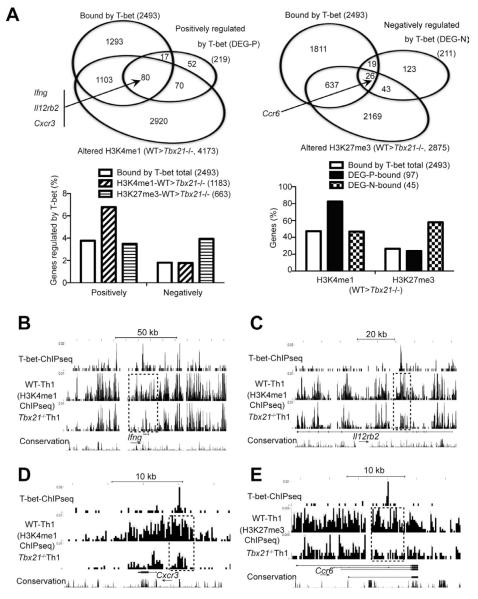
Figure 6. T-bet directly regulates (either positively or negatively) many key molecules of Th1 and Th17 cells.
Naïve CD4+ T cells from WT and Tbx21−/− TBGR mice were cultured under Th1 cell-polarizing conditions for 3-4 days. After resting in IL-2-containing medium for 2 days, cells were sorted for ZsG+ and further expanded under Th1 cell-polarizing conditions for another 3-4 days. Anti-T-bet and histone ChIPseq as well as RNAseq were carried out.
(A) The Venn diagrams show the overlap of genes that are bound by T-bet, positively (upper left) or negatively (upper right) regulated by T-bet at a transcriptional or epigenetic level in Th1 cells. The association of histone modifications and gene regulation were determined in sub-groups of the genes that are bound by T-bet (lower panels). DEG-P and DEG-N stand for differentially expressed genes (DEG) that are regulated by T-bet positively (P) or negatively (N).
(B-E) UCSC genome browser view of T-bet binding, H3K4me1 or H3K27me3 modifications at the Ifng (B), Il12rb2 (C), Cxcr3 (D) and Ccr6 (E) locus are shown. Dotted boxes indicate differences in epigenetic modifications between WT and Tbx21−/− cells around the T-bet binding sites. Arrows indicate the direction of the genes.