Abstract
Objectives
The Korean government has enacted two laws, namely, the Personal Information Protection Act and the Bioethics and Safety Act to prevent the unauthorized use of medical information. To protect patients' privacy by complying with governmental regulations and improve the convenience of research, Asan Medical Center has been developing a de-identification system for biomedical research.
Methods
We reviewed Korean regulations to define the scope of the de-identification methods and well-known previous biomedical research platforms to extract the functionalities of the systems. Based on these review results, we implemented necessary programs based on the Asan Medical Center Information System framework which was built using the Microsoft. NET Framework and C#.
Results
The developed de-identification system comprises three main components: a de-identification tool, a search tool, and a chart review tool. The de-identification tool can substitute a randomly assigned research ID for a hospital patient ID, remove the identifiers in the structured format, and mask them in the unstructured format, i.e., texts. This tool achieved 98.14% precision and 97.39% recall for 6,520 clinical notes. The search tool can find the number of patients which satisfies given search criteria. The chart review tool can provide de-identified patient's clinical data for review purposes.
Conclusions
We found that a clinical data warehouse was essential for successful implementation of the de-identification system, and this system should be tightly linked to an electronic Institutional Review Board system for easy operation of honest brokers. Additionally, we found that a secure cloud environment could be adopted to protect patients' privacy more thoroughly.
Keywords: Access to Information, Information Systems, Research Design, Research Ethics, Biomedical Research
I. Introduction
As Electronic Medical Record (EMR) systems have been widely adopted, research using EMR data has been widespread due to its ease of accessing large amounts of clinical data. Therefore, concerns regarding the privacy and security of patients' medical records have been highlighted. In the United States, the Health Insurance Portability and Accountability Act (HIPAA) defined guidelines for the secondary use of medical records. Based on the HIPAA, the Office for Civil Rights published a guideline for de-identification of medical records recently [1]. Korean government also enacted two laws, namely, the Personal Information Protection Act [2] and the Bioethics and Safety Act [3], to prevent the unauthorized use of medical information. In particular, the revised Bioethics and Safety Act extends its scope from embryo research and genetic research to general biotechnology research to prevent any infringement of human dignity. Therefore, the revised act applies to both prospective studies and retrospective studies. However, it is almost impossible to obtain each participant's informed consent for most cases of retrospective studies. Therefore, Korean governmental regulations also suggest the de-identification of personal health information as an alternative.
To protect patients' privacy by complying with the governmental regulations and improve the convenience of research, Asan Medical Center (AMC) has been developing a biomedical research platform. Based on thorough review of the two Korean regulations and well-known previous biomedical research platforms which support de-identification, we implemented the prototype of a de-identification system. The de-identification system proposed in this paper includes not only identifier removal methods but also the necessary platform, such as user client programs and the interfaces for other programs. Since AMC is the biggest hospital in Korea and it has actively adopted health information technology to improve the quality of care and to make a clinical workflow more efficient [4], our experience and lessons learned from developing the de-identification system will be helpful to other hospitals.
II. Methods
We first reviewed the Personal Information Protection Act and the Bioethics and Safety Act to define the scope of the de-identification methods. Then, we also investigated representative previous biomedical research platforms, such as the Stanford Translational Research Integrated Database Environment (STRIDE) [5], Informatics for Integrating Biology and the Bedside (i2b2) [6], and the Research Patient Data Registry (RPDR) [7] since de-identification will serve as a part of the research platform.
1. Review of Regulations
The scope of de-identification for biomedical research in Korea can be categorized into three parts as shown in Table 1. First, we have to encrypt sensitive data such as the Korean resident registration number, which is similar to the Social Security number in the United States, when we store those data in a database system. Since the Korean resident registration number is a unique life-long personal identifier and is used as a unique key to distinguish a specific person, this number must be encrypted securely. Also, the Institutional Review Board (IRB) process should be tightly linked to the de-identification process. Second, protected health information (PHI) must be removed if researchers do not have the proper informed consent. We have to de-identify not only all direct identifiers but also all possible quasi-identifiers. Quasi-identifiers are values of variables within a dataset that are not unique but might be empirically specific by combining them. Last, we should establish a bio-bank to manage human materials. In this paper, we will focus on the de-identification of medical information.
Table 1.
Scope of de-identification in Korean regulations
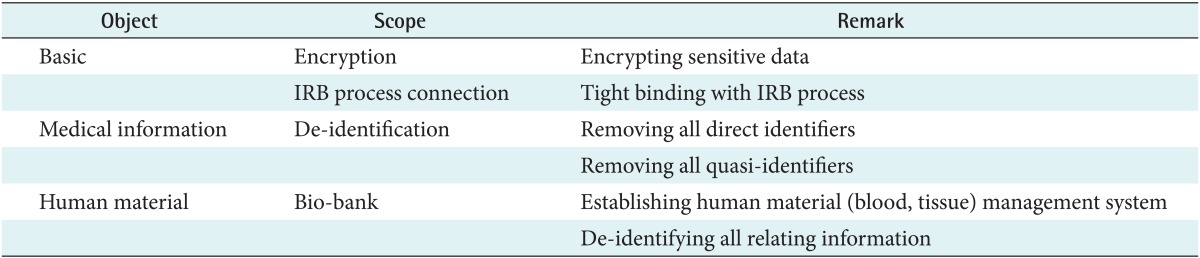
IRB: International Review Board.
2. De-identification System Design
STRIDE is a standard-based informatics platform supporting clinical and translational research [5,8]. It comprises three main databases, namely, a clinical data warehouse (CDW), a bio-specimen database, and research databases. Above those databases, an anonymous cohort identification tool, a patient cohort data review tool, clinical data extraction, research data management, and bio-specimen data management are served. STRIDE follows the HIPAA rules for de-identification. i2b2 integrates data from multiple sources, combines research data with clinical data, and focuses on cohorts and patient populations without PHIs [6,9]. RPDR is a centralized clinical data registry. Researchers access this data using the RPDR online query tool with user-defined queries to extract the aggregate numbers of patients and, with proper IRB approval, obtain detailed clinical data. RPDR secures patient information by controlling and auditing the distribution of patient data within the guidelines of the IRB and with the use of several built-in, automated security measures [7,10].
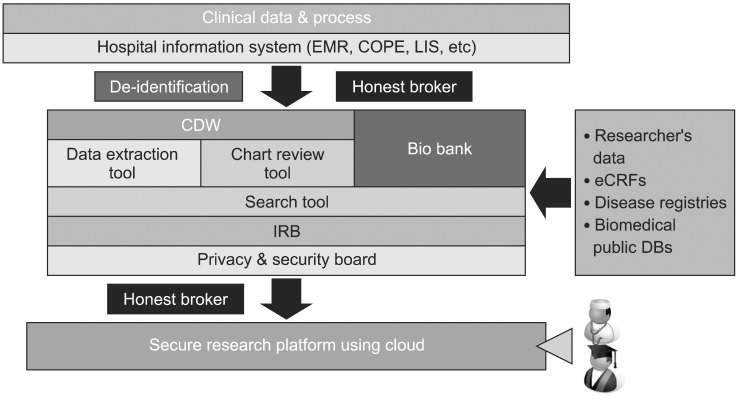
Based on the defined scopes and review of other systems, we designed an overall de-identification system and related processes as shown in Figure 1. Physicians, nurses, or other medical staff members generate the clinical data using the hospital information system, including EMR, computerized physician order entry (CPOE), or laboratory information management system. In this step, all data should be identifiable for proper patients' care. The de-identification tool removes the structured identifiers, such as, names, telephone numbers, and patient IDs. It also masks the identifiers in the unstructured data, i.e., names in the text, using regular expressions. Only honest brokers which are humans or a system can reverse this process. The de-identified data and identifiable data are stored in CDW and reorganized for easy search and analysis. Also, research data from clinical trials (electronic case report forms), disease registries, or the researcher's own data which stored as Microsoft Access or Excel format can be transferred in CDW. If necessary, public biomedical databases may be linked into CDW. Users can access this system using two clients, such as the search tool and the chart review tool. The search tool should have a user-friendly interface and support ad-hoc queries, since researchers want to check the size of the possible research cohort which satisfies the necessary conditions. Users can review the de-identified clinical data using the chart review tool. The de-identified clinical data should include all possible medical records related to each patient, including disease names or codes, operation names or codes, laboratory results, medication, and progress notes. For stricter protection, privilege should be checked and maintained by IRB or other an internal privacy board regularly. If users need the identifiable data or extract data, they should contact honest brokers with the proper informed consents and IRB approval.
Figure 1.
Diagram of Asan Medical Center de-identification system. EMR: Electronic Medical Record, COPE: computerized physician order entry, LIS: laboratory information system, CDW: clinical data warehouse, IRB: International Review Board, eCRF: electronic case report form, DB: database.
3. System Implementation
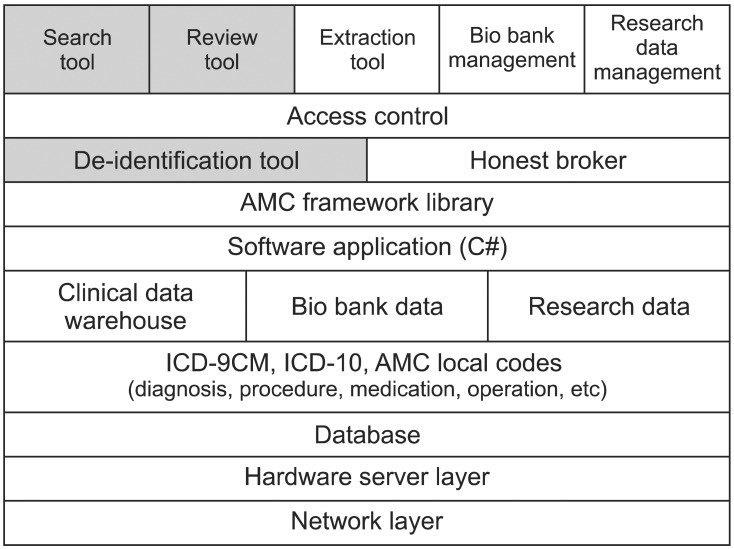
The overall system architecture stack is shown in Figure 2. The de-identification system is based on diverse layers. The network layer, hardware layer, and database are located at the bottom of the stack. The terminology layer is located above them. AMC uses local codes for diagnosis, procedure, medication, operation, and laboratory tests, and AMC maps the necessary codes to the standard terminology, such as the International Classification of Diseases (ICD)-10 and ICD-9-CM. The data are stored in CDW, the bio-bank database, or the research database, respectively. We implemented the necessary programs based on the Asan Medical Center Information System (AMIS) framework. The AMIS framework library was implemented using the Microsoft .NET Framework and C#. We developed all clients based on the AMC enterprise data warehouse (EDW) which has been used for both clinical research and hospital management since 2001. However, we are developing a new dedicated clinical research data warehouse to support researchers more efficiently.
Figure 2.
Overall system architecture of Asan Medical Center (AMC) biomedical research platform. ICD: International Classification of Diseases.
To focus on development of the de-identification, we will introduce only three tools in this paper: the de-identification tool, data review tool, and data search tool. They are indicated in gray in Figure 2.
III. Results
1. Research ID Generation
To reinforce the protection of privacy, we replace patient IDs with research IDs when those patient IDs are requested for the purpose of data review. We use a random number generator to create the candidate research ID for each patient and then check for duplication as shown in Figure 3. Therefore, we can assign a unique randomly generated ID for each patient. Since we will give this research ID to users when they are reviewing de-identified data, we decided not to use a hash function. The hash code is too long and complicated for this purpose. Also, the random number generation function returns 8-digit random numbers to maintain a format similar to that of the patient ID. Only honest brokers can access the mapping table of patient IDs and research IDs.
Figure 3.
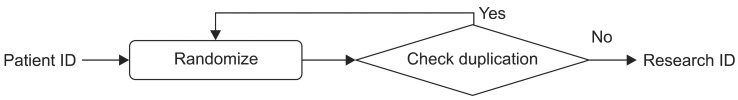
Flowchart for generating Research ID.
2. Direct Identifier Removal
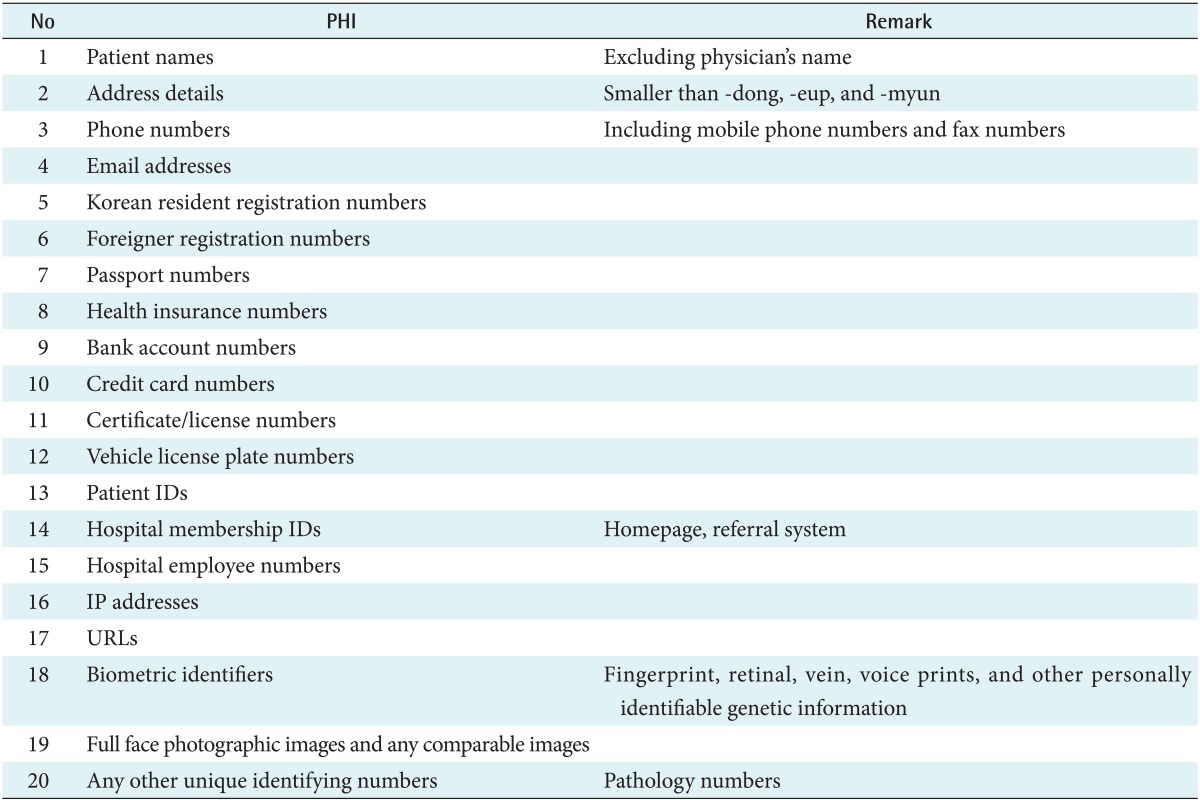
We tried to remove 20 PHIs defined by the AMC Privacy & Security Board as shown in Table 2. Since there is no detailed governmental definition of PHIs in Korea, i.e., HIPAA PHI definition, we defined the institutional PHIs. The major difference between the PHIs of AMC and those of HIPAA is that AMC did not define the date directly related to an individual as PHI. Researchers in AMC strongly insist that the date is necessary for clinical research.
Table 2.
Twenty protected health informations defined by Asan Medical Center
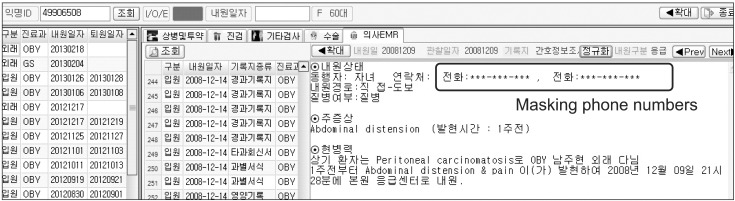
The identifiers in the structured format were easily removed. However, it is complicated to remove the identifiers in the unstructured format such as free texts. There have been several previous works on the automatic de-identification of textual data in EMR [11,12], and some tools have shown reliable performance [13,14]. However, physicians in AMC wrote free texts using Korean as well as English. Therefore, it was hard to apply English-oriented de-identification tools. To overcome this problem, we applied a heuristic approach using regular expressions as a first step [15]. We masked the 20 PHIs in free texts as shown in Figure 4. The developed method was verified by 6,502 carefully chosen clinical notes of 66 types, including inpatient, outpatient, emergency room, and operating room notes. Those clinical notes were written by 498 different physicians. Five human annotators reviewed all of the notes manually to confirm performance of the automatic method. The de-identification tool achieved 98.14% of precision and 97.39% of recall. Here, precision means that the ratio of the correctly masked PHIs versus the masked data and recall represents the ratio of the successfully masked PHIs versus the entire PHIs. There were 1,861 PHIs in the 6,502 clinical notes. Among them, 1,837 PHIs were accurately masked, 18 non-PHIs were removed, and 24 PHIs still remained. When reviewing, we could find only 4 PHIs, which were phone numbers, patient names, addresses, and patient IDs. Other PHIs were not found in free texts.
Figure 4.
The PHIs such as two phone numbers are masked with asterisks in the outpatient progress note.
3. Quasi-Identifier Removal
Since Korean regulations strictly prohibit the use of quasi-identifiers for research purpose, we adopted k-anonymity [16,17]. k-anonymity prevents the identification of a patient when there are less than k similar data. Though there is no standard on deciding k, El Emam [16] and El Emam et al. [17] proposed 5-anonymity in health records by a rule-of-thumb. It means that if there are less than 5 patients' data, anonymity can be guaranteed. Based on this rule-of-thumb, we simply do not provide the results which have less than 5 patients' data to guarantee 5-anonymity in a simply way.
4. Search Tool
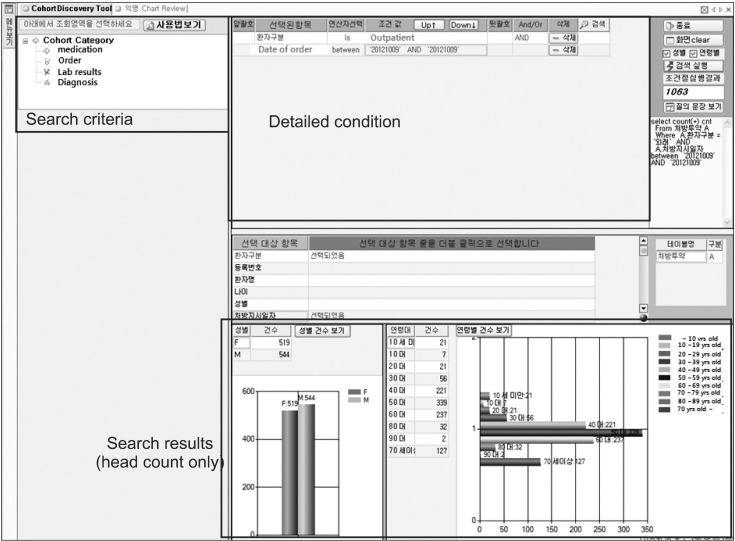
A screenshot of the AMC anonymized search tool is shown in Figure 5. Users can find the number of patients by setting several phenotypes including a diagnosis name/code, an operation name/code, lab results, or medication. A user selects search criteria in the left panel by double-click or drag-and-drop, and sets the detailed conditions of the selected criteria in the upper right panel. Finally, the search results are displayed in the lower panel with graphs. When deciding the detailed conditions, a researcher can use diverse operators, such as 'equal', 'bigger than', 'between', and other necessities. The left graph in the result panel shows the number of patients categorized by sex, and the right one presents the number of patients by age groups. In Korea, ethnic group is not important.
Figure 5.
User interface of search tool. In the left panel, user can choose the search criteria such as medication, order, lab results, and diagnosis. User can set the detailed search parameter in the upper right panel. In this figure, user searched the total number of outpatients in October 9, 2012. The lower right panel shows the search results.
5. De-identified Chart Review Tool
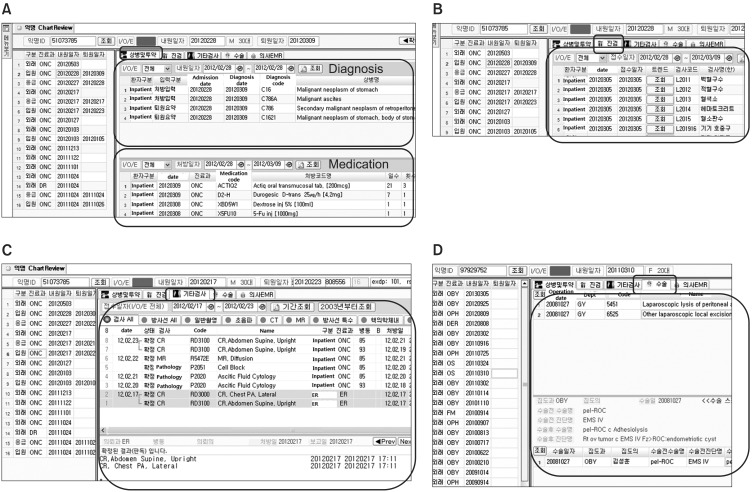
Figure 6 depicts a user interface of the chart review tool. Figure 6A shows the diagnosis and medication tab which integrates AMC local diagnosis codes, ICD-10, and the related codes which were input by physicians. This tab also provides all medication orders with related drug information. The lab result tab, as seen in Figure 6B, shows the individual laboratory results as well as the overall trend of the chosen test. Radiology and pathology reports are also reviewed as in Figure 6C. The operative reports tab, as seen in Figure 6D, provides operation names, operative diagnosis names, and ICD-9-CM codes. The EMR tab displays textual information in progress notes, admission notes, and discharge summary as shown in Figure 4.
Figure 6.
User interface of chart review tool. (A) Diagnosis & medication, (B) lab results, (C) radiology & pathology reports, and (D) operative reports.
Using the given research ID, the user can access the de-identified patient's chart including diagnosis, medication, lab results, radiology and pathology reports, operative reports, progress notes, admission notes, and discharge summary. Thus, researchers can review all medical records related to the chosen patient.
IV. Discussion
We implemented the prototype of a de-identification system designed to comply with Korean governmental regulations. Lessons we learned by implementing the system at the largest medical center in Korea can be summarized as follows. First, we reconfirmed that a data warehouse is essential for the successful implementation of the de-identification system, as most of the previous de-identification systems are based on data warehouses. Without an EDW in AMC, it would be almost impossible to implement the prototype system since the data in legacy systems, such as EMR and CPOE, cannot be de-identified. Second, a clinical research data warehouse is more suitable than the usual EDW. Some hospitals have implemented de-identification systems using EDW, for example, the Ohio State University Medical Center Information Warehouse [18]. However, there are many benefits of having a dedicated clinical research data warehouse for the de-identification system. Researchers usually require a large amount of raw data instead of summarized reports. Also, the de-identification system requires unique tools and processes, i.e., honest broker, de-identified ad-hoc query interface, anonymized chart review, and IRB approval interface, which are not necessary for EDW. Third, an electronic IRB system (e-IRB) is needed, and it must have an interface for easy and automatic transferring of approval or waiver into the de-identification system. If there is only a paper-based IRB system or e-IRB without an interface to the de-identification system, the IRB approval must be checked manually. This is time consuming and inconvenient for researchers. Last, when extracting identifiable data with IRB approval, a digital rights management software or a secure private cloud is necessary to prevent data breach caused by hacking or carelessness of researchers. Data breach makes it meaningless to protect patient's privacy using the de-identification system. Also, a secure cloud system can offer powerful computing resources to handle big data such as genomic data.
Acknowledgments
This study was supported by a grant (2012-543) from the Asan Institute for Life Sciences, Seoul, Korea.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Office for Civil Rights. Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule [Internet] Washington (DC): US Department of Health & Human Service; c2013. [cited at 2013 Mar 28]. Available from: http://www.hhs.gov/ocr/privacy/hipaa/understanding/coveredentities/De-identification/hhs_deid_guidance.pdf. [Google Scholar]
- 2.Korea Ministry of Government Legislation. The Personal Information Protection Act [Internet] Seoul, Korea: Korea Ministry of Government Legislation; c2013. [cited at 2013 Mar 28]. Available from: http://www.law.go.kr/lsEfInfoP.do?lsiSeq=136728#0000. [Google Scholar]
- 3.Korea Ministry of Government Legislation. The Bioethics and Safety Act [Internet] Seoul, Korea: Korea Ministry of Government Legislation; c2013. [cited at 2013 Mar 28]. Available from: http://www.law.go.kr/lsEfInfoP.do?lsiSeq=137037#0000. [Google Scholar]
- 4.Ryu HJ, Kim WS, Lee JH, Min SW, Kim SJ, Lee YS, et al. Asan medical information system for healthcare quality improvement. Healthc Inform Res. 2010;16(3):191–197. doi: 10.4258/hir.2010.16.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE: an integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009;2009:391–395. [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalichowski R, Keogh D, Chueh HC, Murphy SN. Calculating the benefits of a Research Patient Data Repository. AMIA Annu Symp Proc. 2006:1044. [PMC free article] [PubMed] [Google Scholar]
- 8.Stanford Center for Clinical Informatics, Stanford School of Medicine. Stanford Translational Research Integrated Database Environment (STRIDE) [Internet] Stanford (CA): Stanford School of Medicine; c2013. [cited at 2013 Mar 28]. Available from: https://clinicalinformatics.stanford.edu/research/stride.html. [Google Scholar]
- 9.National Center for Biomedical Computing. Informatics for Integrating Biology & the Bedside [Internet] Bethesda (MD): National Institutes of Health; c2013. [cited at 2013 Mar 28]. Available from: https://www.i2b2.org/ [Google Scholar]
- 10.Partners Healthcare. Research Patient Data Registry (RPDR) [Internet] Boston (MA): Partners HealthCare; c2013. [cited at 2013 March 28]. Available from: http://rc.partners.org/rpdr. [Google Scholar]
- 11.El Emam K. Methods for the de-identification of electronic health records for genomic research. Genome Med. 2011;3(4):25. doi: 10.1186/gm239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser R, Willison D. Tools for de-identification of personal health information. [Toronto]: Pan Canadian Health Information Privacy Group; 2009. [cited at 2013 Apr 3]. Available from: https://www.infoway-inforoute.ca/index.php/resources/video-gallery/doc_download/624-tools-for-de-identification-of-pseronal-health-information. [Google Scholar]
- 13.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleger L, Molnar K, Savova G, Xia F, Lingren T, Li Q, et al. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform Assoc. 2013;20(1):84–94. doi: 10.1136/amiajnl-2012-001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin SY, Shin Y, Choi HJ, Park J, Lyu LM, Kim WS, et al. Deidentification method for bilingual EMR free texts; Proceedings of the 14th World Congress on Medical and Health Informatics; 2013 Aug 20-23; Copenhagen, Denmark. [Google Scholar]
- 16.El Emam K. Heuristics for de-identifying health data. IEEE Secur Priv. 2008;6(4):58–61. [Google Scholar]
- 17.El Emam K, Dankar FK, Issa R, Jonker E, Amyot D, Cogo E, et al. A globally optimal k-anonymity method for the de-identification of health data. J Am Med Inform Assoc. 2009;16(5):670–682. doi: 10.1197/jamia.M3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Erdal S, Silvey SA, Ding J, Riedel JD, Marsh CB, et al. Toward a fully de-identified biomedical information warehouse. AMIA Annu Symp Proc. 2009;2009:370–374. [PMC free article] [PubMed] [Google Scholar]