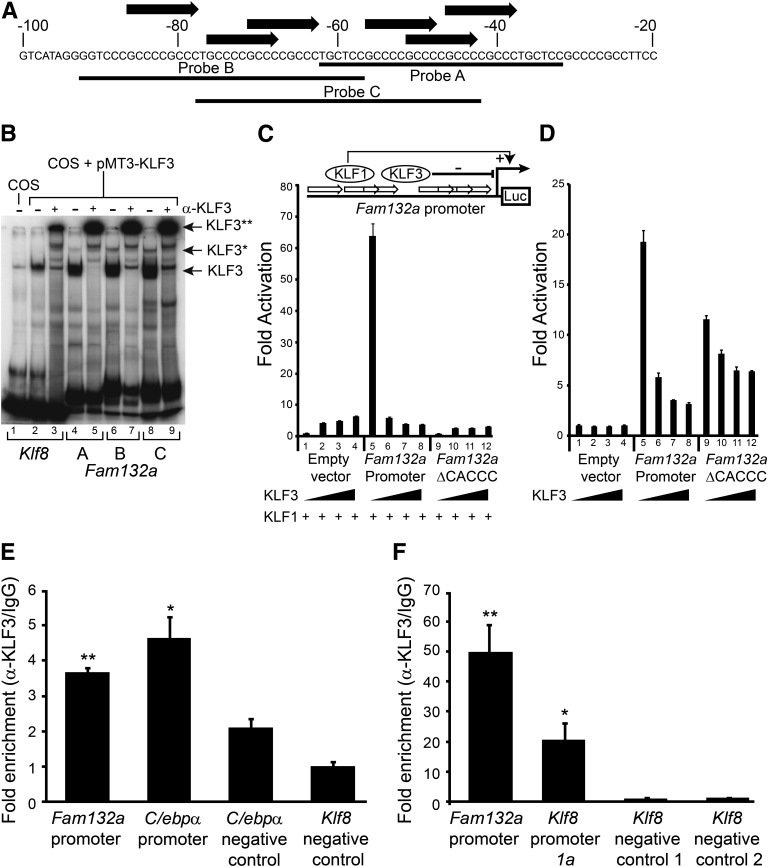
FIG. 5.
KLF3 binds the Fam132a promoter in vitro and in vivo and is able to repress its activity. A: Probe design to assess binding of KLF3 to the Fam132a proximal promoter by EMSA: three probes (probes A, B, and C) were designed to provide coverage of major CACCC boxes identified in the promoter. Black arrows indicate CACCC boxes; numbering is with respect to the transcriptional start site. B: EMSA; KLF3 was expressed in COS-7 cells and binding of nuclear extracts to probe A (lanes 4 and 5), probe B (lanes 6 and 7), and probe C (lanes 8 and 9) assessed. Also shown is binding of KLF3 to a previously validated consensus sequence in the Klf8 promoter (29) (lanes 2 and 3). Binding of untransfected COS nuclear extract to the Klf8 probe is shown in lane 1. α-KLF3 indicates an anti-KLF3 antibody used in supershift to validate KLF3 specific binding. Arrows show the positions of probe complexes bound by KLF3. *KLF3, higher-order multimeric complexes. **KLF3, supershift complexes. C: SL-2 cells were transiently transfected with either pGL4.10[luc2], pGL4.10[luc2] containing a region of the wild-type Fam132a proximal promoter, or pGL4.10[luc2] containing a mutant version of the promoter lacking major CACCC boxes (Fam132aΔCACCC). pPac-KLF1 (250 ng) was used to drive activation of the reporter constructs. Increasing amounts of pPac-KLF3 were included to assess repression: 0 ng (lanes 1, 5, and 9), 25 ng (lanes 2, 6, and 10), 50 ng (lanes 3, 7, and 11), and 250 ng (lanes 4, 8, and 12). Renilla vector pGL4.74[HRluc/TK] (1 μg) was used as a transfection control. Shown is the average of three replicates, with error bars representing SEM. Also included is a cartoon summarizing the luciferase reporter assay strategy. White arrows represent Fam132a promoter CACCC boxes, which can be bound by either KLF1 to drive luciferase activity or by KLF3 to repress expression. Luc, luciferase reporter gene. D: 3T3-L1 cells were transiently transfected with either pGL4.10[luc2], pGL4.10[luc2] containing a region of the wild-type Fam132a proximal promoter, or pGL4.10[luc2] containing a mutant version of the promoter lacking major CACCC boxes (Fam132aΔCACCC). Cells were transfected with increasing amounts of pCDNA3-KLF3 to assess repression: 0 ng (lanes 1, 5, and 9), 50 ng (lanes 2, 6, and 10), 250 ng (lanes 3, 7, and 11), and 500 ng (lanes 4, 8, and 12). Renilla vector pGL4.74[HRluc/TK] (1 μg) was used as a transfection control. Shown is the average of three replicates, with error bars representing SEM. E and F: In vivo binding of KLF3 to the endogenous Fam132a promoter was assessed in 3T3-LI (E) and murine erythroleukemia (F) cells by ChIP. Assays were carried out in duplicate for 3T3-L1 cells and in triplicate for murine erythroleukemia cells. Enrichment was determined by real-time PCR and is relative to input signal and IgG precipitated controls. The lowest signals for anti-KLF3 precipitated chromatin were each set to 1.0. Also shown is KLF3 enrichment at previously reported positive control regions in the Klf8 and C/ebpα promoters and additional negative control regions in the Klf8 and C/ebpα locus, where KLF3 has been shown not to bind (14,29). Error bars show SEM; P values indicate the statistical difference between means for Fam132a, C/ebpα, or Klf8 promoters and negative control region 1 in the Klf8 gene locus. **P < 0.01, *P < 0.05.