Abstract
Precise regulation of β-cell function is crucial for maintaining blood glucose homeostasis. Pax6 is an essential regulator of β-cell–specific factors like insulin and Glut2. Studies in the developing eye suggest that Pax6 interacts with Mitf to regulate pigment cell differentiation. Here, we show that Mitf, like Pax6, is expressed in all pancreatic endocrine cells during mouse postnatal development and in the adult islet. A Mitf loss-of-function mutation results in improved glucose tolerance and enhanced insulin secretion but no increase in β-cell mass in adult mice. Mutant β-cells secrete more insulin in response to glucose than wild-type cells, suggesting that Mitf is involved in regulating β-cell function. In fact, the transcription of genes critical for maintaining glucose homeostasis (insulin and Glut2) and β-cell formation and function (Pax4 and Pax6) is significantly upregulated in Mitf mutant islets. The increased Pax6 expression may cause the improved β-cell function observed in Mitf mutant animals, as it activates insulin and Glut2 transcription. Chromatin immunoprecipitation analysis shows that Mitf binds to Pax4 and Pax6 regulatory regions, suggesting that Mitf represses their transcription in wild-type β-cells. We demonstrate that Mitf directly regulates Pax6 transcription and controls β-cell function.
The islets of Langerhans consist of α-, β-, δ-, ε-, and pancreatic polypeptide (PP) cells, which produce the hormones glucagon, insulin, somatostatin, ghrelin, and PP, respectively. Together these hormones regulate fuel metabolism, with insulin and glucagon being essential for glucose homeostasis. Thus, glucagon secreted from α-cells stimulates the mobilization of glucose through gluconeogenesis and glycogenolysis, while β-cell–secreted insulin promotes glucose storage. Defects in α- and β-cell function play a significant role in the ability of diabetic individuals to maintain glycemic control.
Characterization of the transcription factors regulating insulin and glucagon expression has demonstrated their significance to islet cell function and endocrine cell development. For example, pancreatic and duodenal homeobox 1 (Pdx1) is crucial for pancreas development; pancreatic endodermal progenitors fail to proliferate in the absence of Pdx1, resulting in a small pancreatic rudiment (1). Pdx1 is also critical for mature β-cell activity because deletion of this factor from adult β-cells results in a severe diabetic phenotype, which is at least partially caused by reduced insulin and Glut2 expression (2). Pax6 and NeuroD1 are also required for insulin and Glut2 expression, and loss of these transcription factors affects β-cell function and endocrine cell differentiation (3,4). Significantly, mutations in PDX1 (5), PAX6 (6), and NEUROD1 (7) also cause maturity-onset diabetes of the young in humans.
β-Cell function and glucose responsiveness are established during late embryonic and postnatal development (8). Recently, the MafA and MafB transcription factors have been identified as key regulators of these processes because of their ability to regulate genes essential for endocrine cell function, such as insulin, glucagon, Pdx1, and Glut2 (9–11). Embryonic β-cells initially express MafB, but MafB expression is lost in postnatal β-cells, which express MafA instead (12). The switch between MafB and MafA expression in β-cells is associated with the development of functional β-cells. Recent gene profiling studies have shown that MafB regulates genes required for β-cell function during embryonic β-cell differentiation while the same genes are activated by MafA in adult β-cells (12). These gene expression experiments have shown that microphthalmia transcription factor (Mitf) expression is reduced in MafA and MafB mutant embryonic pancreata (12). Mitf, a basic-helix-loop-helix-leucine-zipper transcription factor, regulates melanogenesis by activating transcription of pigment cell-specific genes in both the skin and retina (13,14). In addition, it can act as a transcriptional repressor (14,15) and controls the expression of cell survival (Bcl2) (16) and cell cycle regulatory (Cdk2 and p16/Ink4a) genes (17,18).
In this study, we show that Mitf is produced in the developing pancreas and adult islets. Mitf loss-of-function mice have lower blood glucose levels than wild-type animals in response to an intraperitoneal glucose challenge but also during nonfasted conditions. Mutant islets secrete more insulin upon exposure to high glucose concentrations, and Mitf mutant animals have higher circulating insulin levels in fasted conditions. Additionally, the expression of genes regulating blood glucose levels (insulin and Glut2) and β-cell formation and function (Pax4 and Pax6) is significantly higher in Mitf mutant than in wild-type islets. Promoter occupancy studies show that Mitf binds to the pancreas-specific Pax4 and Pax6 regulatory regions, which suggests that Mitf directly regulates the transcription of these genes in β-cells and thereby modulates β-cell function.
RESEARCH DESIGN AND METHODS
Mice with a point mutation (C to T at nucleotide 916) in the Mitf gene have previously been characterized (19). The Mitf cloudy eye (Mitfce/ce) mutation results in a lack of the Mitf zipper domain due to the presence of a stop codon between the helix-loop-helix and zipper motif. This leads to the synthesis of a truncated protein incapable of DNA binding (19). Mitfce/ce mice have white coat color and small eyes. Heterozygous and wild-type animals both have black coat color. For distinguishing between the latter, DNA samples were sequenced (forward, ggtccattgtcttgttttatcacag; reverse, gtatccaccctctgccat). Wild-type, heterozygous, and homozygous animals are distinguished by C, C/T, and T at bp 916 of the Mitf gene (http://www.informatics.jax.org/searches/accession_report.cgi?id=mgi:1856532). Wild-type and heterozygous Mitf animals were indistinguishable in physiological and histological studies (data not shown), and therefore the data for them are combined as “wt/het” data. All animal work was approved by a local ethics committee for animal research.
Immunohistochemistry.
Pancreata from E15.5 and E18.5 embryos, postnatal day P0, P7, and P21, and 12 week-old mice were fixed for 2 h at 4°C in 4% paraformaldehyde in PBS, washed with PBS, dehydrated, and embedded in paraffin. Sections (6 μm) were used for immunohistochemical analysis as previously described (9).
Antibodies.
Primary antibodies used were as follows: rabbit anti-Mitf (1:1,000; gift from S. Saule, Institut Curie, Paris), rabbit anti-Mitf (1:1,000; Abcam), guinea pig anti-insulin (1:2,000; Millipore), mouse anti-glucagon (1:2,000; SIGMA), sheep anti-somatostatin (1:2,000; American Research Products), guinea pig anti-PP (1:2,000; Millipore), goat anti-ghrelin (1:500; Santa Cruz), rabbit anti-Glut2 (1:500; Chemicon International), rabbit anti-MafA (1:1,000; Bethyl Laboratories), rabbit anti-Nkx6.1 (1:1,000; β-Cell Biology Consortium), rabbit anti-Pax6 (1:1,000; Covance), mouse anti-Pax6 (1:100; Developmental Hybridoma Bank), goat anti-Pdx1 (1:1,000; β-Cell Biology Consortium), mouse anti-islet 1 (1:100; Developmental Hybridoma Bank), and mouse anti-Synaptophysin (1:100; Abcam). Secondary antibodies used were as follows: (1:500) diluted CY2, CY3, CY5 conjugated anti-mouse, -goat, -guinea pig, -rabbit, and -sheep (Jackson ImmunoResearch). DAPI (Invitrogen) was used for nuclear counterstaining (1:6,000).
Blood glucose level measurements.
Random-fed blood glucose samples were collected at a consistent time of the day (1200–1300 h). Fasted blood glucose levels were measured after a 6-h fasting period (800–1400 h) with a handheld glucometer (OneTouch; Lifescan). Mean difference between wild-type and Mitfce/ce was tested with Student t test.
Intraperitoneal glucose tolerance test and insulin secretion measurements.
Intraperitoneal glucose tolerance test (IPGTT) was performed on 12-week- and 6-month-old Mitfce/ce and wild-type mice after overnight fasting (12 h) using an injection of 2 g glucose/kg body wt. Blood glucose levels were measured using a handheld glucometer (OneTouch; Lifescan). Blood was collected from an incision in the distal part of the tail. Measurements were taken at 0, 5, 15, 30, 60, and 120 min after glucose administration for the IPGTT and at 0, 2, 5, and 15 min for glucose-stimulated insulin secretion studies. For the latter, blood was collected into heparin-coated tubes (Sarstedt) and the serum fractions were analyzed with a Mouse Insulin ELISA (Mercodia) according to the manufacturer's instructions.
Insulin secretion assay.
Islets were isolated by collagenase digestion and handpicked under a stereomicroscope (20). Batches (n = 8) of three islets for each condition were kept in secretion assay buffer (SAB) (114 mmol/L NaCl, 4.7 mmol KCl, 1.2 mmol/L KH2PO4, 1.16 mmol/L MgSO4, 20 mmol/L HEPES, 2.5 mmol/L CaCl2, 25.5 mmol/L NaHCO3, and 0.2% BSA; pH 7.2) containing 2.8 mmol/L glucose for 60 min in an incubator at 37°C. Three islets per well were transferred to a 96-well plate containing 200 μL/well SAB but with the addition of 2.8, 8.3, and 16.7 mmol/L glucose with or without 35 mmol/L KCl. After transfer of all islets, the plate was placed in an incubator at 37°C. At 60 min, a buffer sample was removed for measurement of insulin by ELISA.
Glucagon secretion measurements.
Blood samples from random-fed and 12-h-fasted wild-type and Mitf mutant mice were collected into heparin-coated tubes. Samples were chilled immediately and centrifuged at 4°C. Plasma aliquots were stored at −20°C. Glucagon concentrations were determined using a glucagon radioimmunoassay kit (Millipore). Statistical analysis was conducted using Student t test.
Islet isolation, RNA extraction, RT-PCR, and quantitative PCR.
Islet isolation has previously been described (20). RNA from E18.5 and P7 pancreas and islets of 12 week-old mice was extracted with ToTally RNA (Ambion) or RNeasy mini kit (Qiagen). RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop), and RNA quality was assessed using a Bioanalyzer (Agilent). Only samples with a RNA Integrity Number >7.5 were further analyzed. Quantitative PCR was performed in two steps. First, reverse transcription was carried out with SuperScript III (Invitrogen) using 500 ng RNA. For the quantitative PCR reaction, cDNA was diluted 1:200 and all assays were performed with Fast SYBR Green Master Mix on a StepOnePlus Real-Time PCR instrument (Applied Biosystems). Primer sequences are available upon request. Formation of expected PCR products was confirmed by electrophoresis and melting curve analysis. Gene expression data were normalized to the expression of the internal control gene hypoxanthine-guanine phosphoribosyltransferase (HPRT). Experiments were repeated at least five times—each in triplicate. Raw data from real-time PCR measurements were exported from StepOne Software v2.1 and analyzed in Microsoft Excel. The data are shown as mean expression with SEM. Graphs represent the fold change in comparison with the wild-type control samples, set as 1. Mean difference was tested with Student t test.
Image analysis, quantification, and statistical analysis.
Immunofluorescence images were collected with Zeiss Axioplan 2 imaging (Zeiss, Jena, Germany) in AxioVision Rel 4.9 software. Immunohistochemistry and quantification were performed from at least three adult wild-type and Mitfce/ce pancreata. β-Cell mass was assessed by quantifying the pancreatic and insulin-stained area in sections at a 720-μm interval throughout the whole pancreatic organ. The percentage of endocrine cells expressing ghrelin was assessed by quantifying Pax6 and ghrelin-producing cells at a 96 (E18.5) or 120 (P7) μm interval throughout the whole pancreas. Synaptophysin-expressing cells were measured in every 20th section of adult pancreas and normalized to the pancreatic weight. Mean differences were tested with Student t test; n ≥ 3.
Chromatin immunoprecipitation.
β-Cell–derived β-TC6 cells and α-cell–derived α-TC6 (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% calf serum. Cells were transfected with FLAG-tagged Mitf expression plasmid (True ORF cDNA clone MR227086; ORIGene) using Metafectene Pro (Biontex). Cells were harvested 48 h after transfection, fixed in 1% formaldehyde in Dulbecco’s modified Eagle’s medium without serum, and treated with lysis buffer (10 mmol/L Tris-HCl, pH 8.0; 10 mmol/L NaCl; 3 mmol/L MgCl2; 1% NP-40; 1% SDS; and 0.5% sodium deoxycholate [DOC]) and sonicated four times for 5 min on high (30 s on and 30 s off) settings in a Bioruptor sonicator (Diagenode). Protein/DNA chromatin fragments were immunoprecipitated with mouse anti-FLAG antibody (Sigma) or mouse IgG (Santa Cruz) as previously described (21).
Nonquantitative PCR reactions were performed using purified immunoprecipitated DNA with 0.8 μmol/L primers specific for Pax6EE (AGGCACGTCCTGGATGTTAG and CCCCAACCTCATTCTTTTCA), Pax6PE (ACTCAGGCCTGTGGTTATGC and TCAAGAGCGAAGCTGGAAAC), Pax4 (AGGGACAATTAGCCCCAAAC and ACAGAAGCTTTCGACCCAGA), Runx2 (GTTGTTTGATTTGTTTTGAAGG and TTTTACCTAAAATGTGGTTTTTG), and PEPCK (GAGTGACACCTCACAGCTGTGG and GGCAGGCCTTTGGATCATAGCC) transcriptional control region primers. PCR products were visualized on a 1.5% agarose gel in 1× Tris-Borate-EDTA buffer. Each chromatin immunoprecipitation (ChIP) experiment was repeated at least three times using independent chromatin preparations.
Transfection assays.
The Pax6 P0 sequence (−3.8 to 0.2 kb) was PCR amplified from mouse genomic DNA and inserted upstream of a promoterless firefly luciferase plasmid (pGL2basic; Promega). Human embryonic kidney (HEK)293 cells were transfected with pPax6P0-LUC, cytomegalovirus-driven Mitf, and/or Pax6, and Renilla luciferase plasmids (phRL-CMV; Promega) using Metafectene Pro reagent (Biontex). Extracts were prepared 40–48 h later and analyzed for firefly and Renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Mean difference was tested with one-way ANOVA; n = 3.
RESULTS
Mitf is specifically expressed in endocrine cells in the postnatal and adult pancreas.
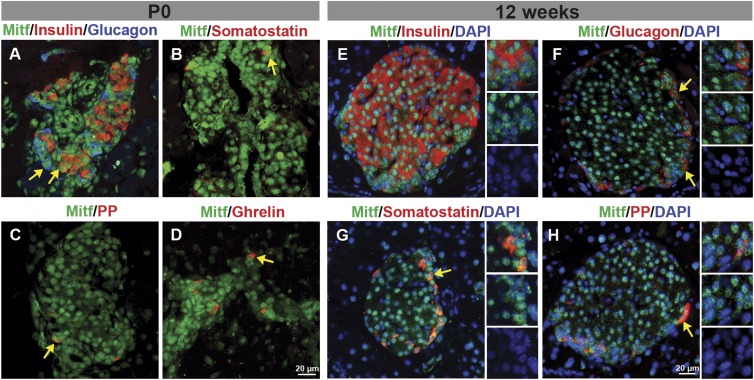
Immunohistochemical analysis was performed to determine which cell types produce Mitf during pancreas development and in the adult. Mitf expression was not detected at E15.5 (data not shown) but was initially observed at E18.5 (Supplementary Fig. 1). At P0, Mitf was found in nuclei of endocrine, exocrine, and ductal cells (Fig. 1A–D and Supplementary Fig. 1). Postnatally, Mitf expression becomes restricted to islet endocrine cells (Supplementary Fig. 1). Analysis of adult wild-type pancreata showed that Mitf is expressed in all hormone-producing cells (Fig. 1E–H). Mitf expression is restricted to nuclei of insulin-producing cells, whereas it is also detected in the cytoplasm of ∼20% glucagon cells (Supplementary Fig. 2).
FIG. 1.
Mitf is found in all five hormone-expressing cell types in the pancreas. A–D: Mitf (green) is detected in the nuclei of hormone-expressing (red) cells at P0. E–H: Mitf expression was observed in insulin-, glucagon-, somatostatin-, and PP-producing cells in islets of 12-week-old mice. Arrows denote Mitf hormone–positive cells.
Mitfce/ce mice have improved blood glucose clearance and elevated insulin secretion.
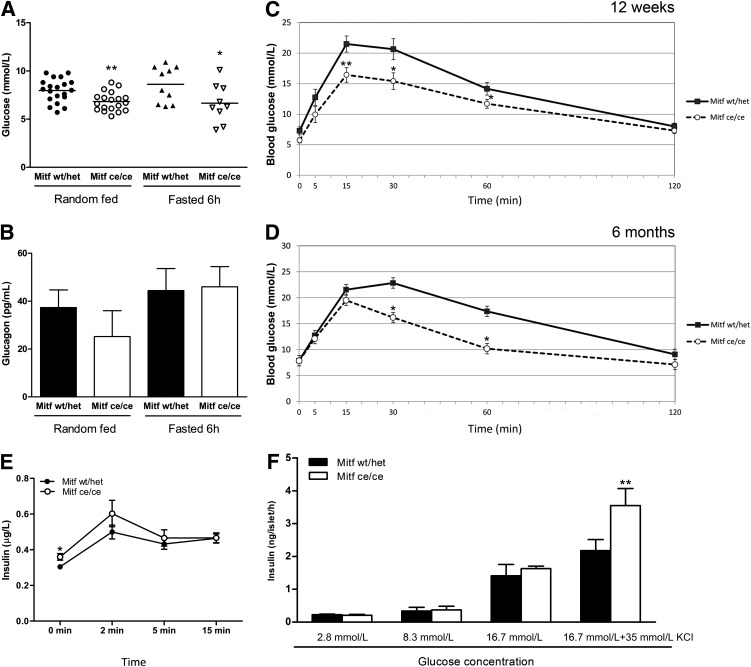
Blood glucose measurements showed that Mitfce/ce mice had significantly lower blood glucose levels than their wild-type littermates whether they were random-fed or fasted (Fig. 2A). In addition, Mitfce/ce animals had improved blood glucose clearance after glucose injection at 12 weeks and 6 months. Blood glucose levels stayed below 20 mmol/L even at 15–30 min after the glucose injection (Fig. 2C and D) and returned faster to homeostatic conditions. Glucose-stimulated serum insulin levels were measured to determine whether this improved glucose clearance is a direct result of altered β-cell function. Interestingly, Mitf mutant animals have significantly higher serum insulin levels in fasting conditions than wild-type animals (0 min [Fig. 2E]), which suggests that Mitf is required for proper β-cell function. In contrast, plasma glucagon levels were not significantly changed in Mitfce/ce animals compared with wild-type littermates in both fasted and random fed conditions (Fig. 2B). Stimulation of Mitfce/ce and wild-type isolated islets with 16.7 mmol/L glucose and 35 mmol/L KCl resulted in a 60% increased insulin secretory response (Fig. 2F). Our results demonstrate that Mitf mutant mice have enhanced glucose tolerance and are protected from high blood glucose levels by elevated insulin secretion.
FIG. 2.
Improved glucose tolerance and increased insulin secretion in Mitfce/ce mice. A: Blood glucose levels from random-fed and 6-h-fasted wild-type (wt/het) and Mitf mutant mice, n ≥ 9. B: Plasma glucagon levels from random-fed and 6-h-fasted wild-type and Mitfce/ce animals, n ≥ 3. C and D: IPGTT (with overnight, 12-h fasting) of 12-week (C) and 6-month-old (D) Mitf mutant and wild-type mice. Blood glucose measurements were taken at 0, 5, 15, 30, 60, and 120 min after the intraperitoneal glucose injection (2 g of D(+) glucose/kg body wt). E: In vivo glucose-stimulated insulin secretion: intraperitoneal glucose injection with 2 g D(+) glucose/kg body wt. Insulin levels were measured 0, 2, 5, and 15 min after injection; n ≥ 10. t test, *P value <0.05, **P value < 0.01. F: Insulin secretion profile of Mitfce/ce mice. Islets isolated from Mitfce/ce and control mice treated with different glucose concentrations (2.8, 8.3, and 16.7 mmol/L glucose and 16.7 mmol/L glucose plus 35 mmol/L KCl). Insulin levels were assessed with insulin ELISA; n ≥ 3. **P value < 0.01 with two-way ANOVA.
Number of endocrine cells is unchanged in Mitfce/ce animals.
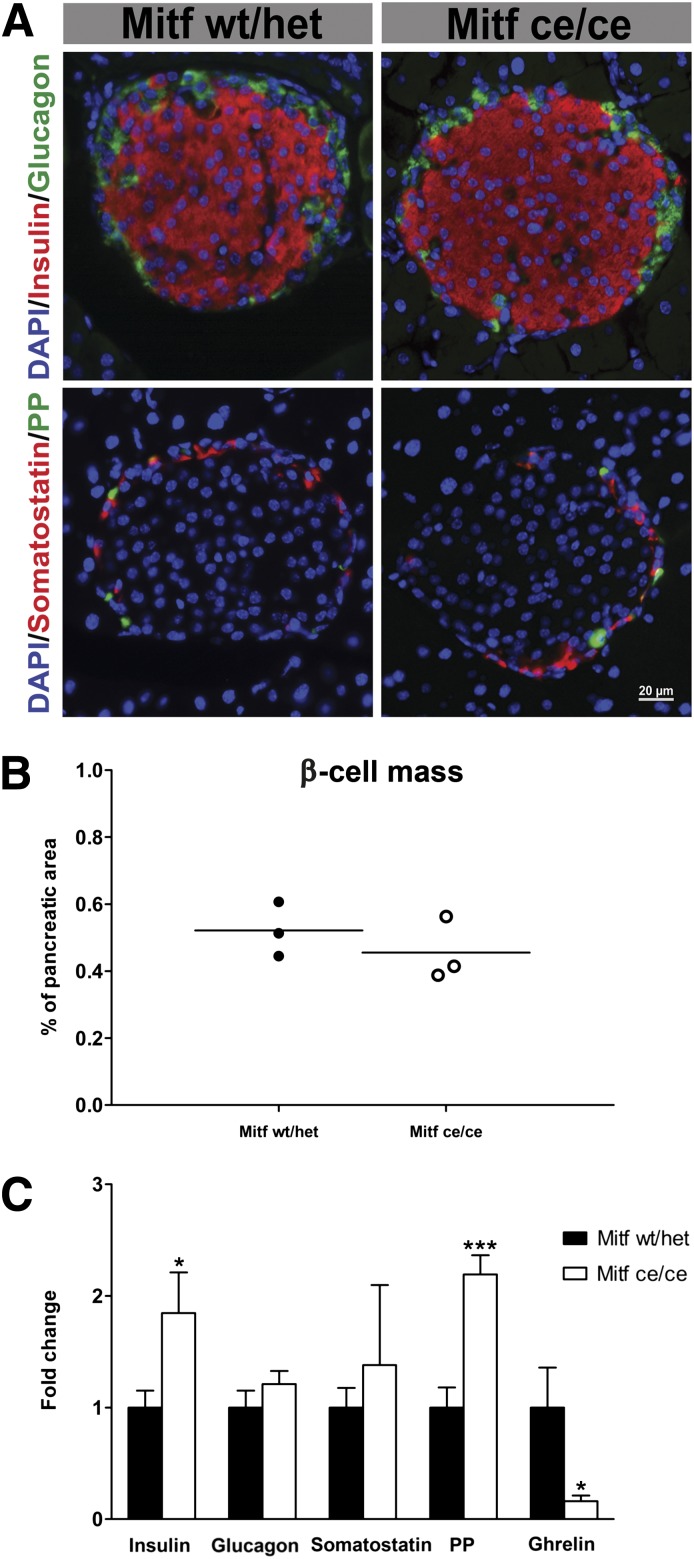
Immunohistochemical analysis of wild-type and Mitfce/ce pancreatic sections showed no obvious changes in the appearance of pancreatic islets (Fig. 3A). For determination of whether the elevated serum insulin levels observed in Mitfce/ce mice result from an increase in β-cell mass, quantitative immunohistochemical analysis was performed. The percentage of insulin-labeled wild-type and Mitfce/ce cell area and total islet cell area were comparable (Fig. 3B and Supplementary Fig. 4C and D) suggesting that Mitf is not required for β-cell specification or regulation of β-cell mass. Insulin and PP mRNA levels were significantly increased in Mitfce/ce islets (Fig. 3C), while ghrelin transcription was decreased to only 20% of wild-type (Fig. 3C). Immunohistochemical quantification of ghrelin-producing cells revealed no differences in the percentage of ghrelin cells within the endocrine Pax6+ cell population at E18.5 and P7 (Supplementary Fig. 3A–E), while ghrelin mRNA levels were already reduced by P7 (Supplementary Fig. 3F). In accordance with previous reports (22) ghrelin cells were almost absent in adult wild-type and Mitfce/ce islets, which precluded a quantitative analysis at this time point. Quantitative immunohistochemical analysis showed no significant increase in PP cell number (data not shown) in Mitfce/ce islets. These findings suggest that insulin, ghrelin, and PP production is altered in individual hormone-expressing cells. Mitfce/ce and wild-type β-cells were analyzed using electron microscopy to determine whether the increase in insulin mRNA levels results in enhanced production of insulin secretory granules. Mitfce/ce β-cells have the same number of insulin granules per cell, and granule morphology is comparable with the wild-type (Supplementary Table 1 and Supplementary Fig. 4A and B). Our findings illustrate that Mitf is critical for proper hormone expression but not essential for endocrine cell specification and maintenance.
FIG. 3.
Wild-type (wt/het) and Mitfce/ce mutant islets consist of insulin, glucagon, somatostatin, and PP cells (A). B: Average β-cell area is unchanged in Mitfce/ce animals; n = 3. C: Quantitative RT-PCR measurement of pancreatic hormone levels in Mitf mutant and wild-type islets (n > 5 per genotype.) The data were normalized to HPRT mRNA levels and are presented as relative to control (set as 1). t test, *P < 0.05, ***P < 0.001.
Gene expression of key β-cell genes is increased in Mitfce/ce islets.
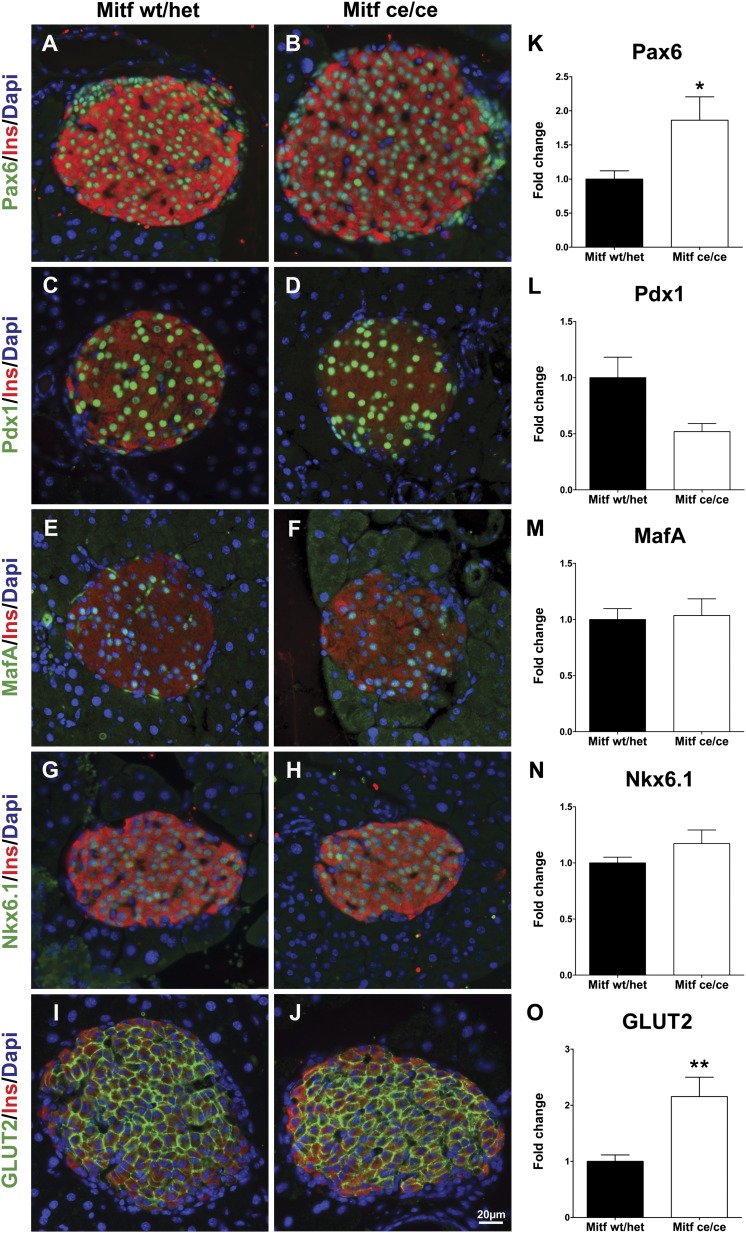
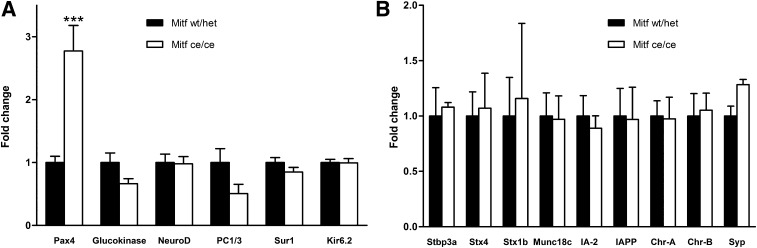
Gene expression analysis and immunohistochemistry were performed to further characterize Mitfce/ce islets. First, we analyzed the expression level and distribution of transcription factors involved in β-cell development and/or function. The cellular distribution of known β-cell factors Pax6, Pdx1, MafA, and Nkx6.1 was unchanged (Fig. 4A–H). However, a twofold increase in Pax6 and Pax4 mRNA levels was observed in Mitfce/ce islet samples (Figs. 4K and 5). Interestingly, β-Cell–specific expression of Glut2, a known Pax6 target gene (3) was increased twofold in mutant islets (Fig. 4O), while the transcription of other glucose-sensing, metabolizing, and insulin secretory genes was unaffected (Fig. 5). These results suggest that Mitf is controlling β-cell function by directly regulating genes required for β-cell function and differentiation.
FIG. 4.
Twelve-week-old Mitfce/ce mice show no obvious differences in the expression pattern of markers characteristic for functional β-cells when analyzed by immunohistochemistry. A and B: Pax6 (green) and insulin (red). C and D: Pdx1 (green) and insulin (red). E and F: MafA (green) and insulin (red). G and H: Nkx6.1 (green) and insulin (red). I and J: GLUT2 (green) and insulin (red). K–O: Quantitative RT-PCR analysis of expression of Pax6, Pdx1, MafA, Nkx6.1, and GLUT2 in mutant (Mitfce/ce) and wild-type (wt/het) islets; n > 5 per genotype. t test, *P < 0.05, **P < 0.001. INS, insulin.
FIG. 5.
Quantitative gene expression profile of pancreatic transcription factors and glucose metabolism–related (A) and insulin secretory (B) genes in Mitf mutant (Mitfce/ce) and wild-type (wt/het) islets (n ≥ 4 per genotype). The data were normalized to HPRT mRNA and are presented relative to control (set to 1). IA-2, insulinoma-associated protein-2; IAPP, islet amyloid polypeptide. ***P < 0.001.
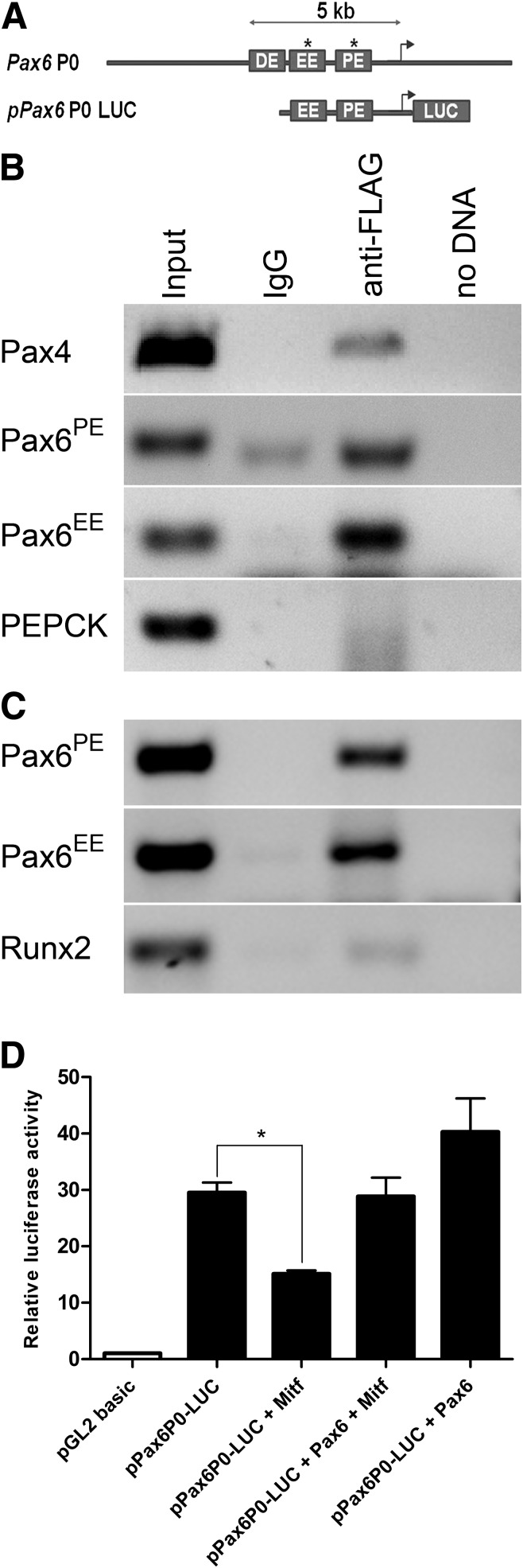
Mitf binds to the pancreas-specific regulatory sequences of the Pax4 and Pax6 genes.
Previous studies have established that Mitf acts as a transcriptional activator and repressor in the developing eye (13–15). We hypothesize that Mitf directly controls Pax6 transcription, since Pax6 mRNA levels were upregulated in Mitfce/ce mice (Fig. 4K) and Mitf overexpression prevents basic fibroblast growth factor (bFGF)-induced Pax6 upregulation in cultured retinal pigmented epithelial cells (14,23). The mouse Pax6 gene contains several regulatory elements that confer tissue-specific expression in the pancreas, eye, and brain (24). Among those, the P0 proximal regulatory element (PE) is required for embryonic pancreas-specific Pax6 expression at physiological levels (25), while ectodermal expression is controlled by the EE element (EE). ChIP analysis was performed in mouse α-TC6 and β-TC6 cells transfected with FLAG-tagged Mitf to determine whether Mitf occupied the Pax6 EE and PE sequences (Pax6EE and Pax6PE). An enrichment of both Pax6EE and Pax6PE was found using primer pairs that span the Pax6PE and Pax6EE regions compared with control sequences only after α-FLAG immunoprecipitation (Fig. 6B and C). A similar analysis demonstrated that flag-tagged Mitf also immunoprecipitates Pax4 regulatory elements in β-TC6 cells (Fig. 6B). For further assessment of functional regulation of the Pax6 P0 promoter, Pax6 and Mitf were cotransfected with a Pax6P0-driven reporter construct in HEK293 cells (Fig. 6A and D). Pax6P0 firefly luciferase (pPax6P0-LUC) activity was enhanced upon cotransfection with Pax6, while cotransfection with Mitf resulted in transcriptional repression. Cotransfection of Mitf and Pax6 resulted in loss of Mitf-mediated repression, suggesting that interactions between these factors are critical for the transcriptional control of the Pax6P0 regulatory region. The ChIP and luciferase reporter data are complimentary and demonstrate that Mitf directly binds to and regulates Pax6 regulatory elements, suggesting that increased Pax6 mRNA levels in Mitfce/ce are a direct effect of the loss of Mitf.
FIG. 6.
Mitf binds to and regulates Pax6 expression in β-TC6 cells. Pancreas-specific Pax6 expression is partially regulated by upstream regulatory elements (summarized in ref. 25). A: The pPax6P0-LUC construct contains the EE and PE elements. B: ChIP analysis was performed on β-TC6 cells transfected with an expression vector containing FLAG-tagged Mitf. The anti-FLAG precipitated chromatin was analyzed by nonquantitative PCR using primers specific for Pax4, Pax6EE, Pax6PE, and PEPCK control region sequences. C: ChIP analysis of α-TC6 cells transfected with FLAG-tagged Mitf. PCR analysis with Pax4, Pax6EE, Pax6PE, and Runx2 control region sequences. Control PCR reactions were run with input chromatin (1:100 dilution), and DNA was obtained after precipitation with mouse IgG or no DNA. ChIP analysis was repeated from at least three independent chromatin preparations. D: Dual luciferase reporter assays show that pPax6P0-LUC activity is enhanced by cotransfection with Pax6 in HEK293 cells but repressed by cotransfection with Mitf.
DISCUSSION
Previous studies have established Mitf as a master regulator of pigment cell differentiation (rev. in 26). Our previous results have shown that Mitf expression is reduced in MafA and MafB mutant embryonic pancreata (12) suggesting that Mitf may be involved in β-cell differentiation and function. In this study, we demonstrate that Mitf is initially expressed in the pancreatic epithelium at E18.5 and that its expression becomes restricted to adult islet cells. Mitf inactivation results in altered β-cell function, due to an enhanced insulin secretory response, without changes in β-cell mass. In contrast, the transcription of genes critical for the regulation of blood glucose levels (insulin and Glut2) and β-cell formation and function (Pax4 and Pax6) was significantly upregulated in Mitfce/ce islets, suggesting that Mitf is critical for adult β-cell function.
During development, Mitf was initially detected in all pancreatic epithelial cells from E18.5 onward (Supplementary Fig. 1). Interestingly, Mitf expression increases and becomes progressively restricted to islet cells postnatally and is not detected in ductal and exocrine cells after the first weeks of birth. Our findings are supported by a gene-profiling study of Ngn3+ endocrine progenitor cells and their descendants, which showed that Mitf transcription increases postnatally, with the highest expression observed in adult islet cells (27). The late onset of Mitf expression and localization in endocrine cells coincides with profound morphological and physiological changes in the pancreas: endocrine cells cluster into mature islets, with β-cells in the core and other hormone-producing cells at the periphery. In addition, and perhaps most importantly, the glucose responsiveness of β-cells is established at this time (8). During specification of zebrafish melanocytes, Mitf is specifically required for late steps of differentiation (28), which also supports a role for Mitf in β-cell maturation or function.
To determine Mitf’s function in the developing and adult pancreas, we analyzed a Mitf mutant mouse model. Mitfce/ce pancreata were histologically indistinguishable from wild-type littermates, β-cell area was comparable, and no changes in adult islet cell composition were observed. However, as evident from our in vitro studies on isolated islets, the insulin response to high glucose and KCl concentrations was significantly greater in Mitfce/ce animals than in wild-type, whereas insulin response to a stimulation with lower glucose concentrations was not significantly affected (Fig. 2F). This shows that the total β-cell secretory capacity is increased in mutant mice. Interestingly, Mitfce/ce animals also had fasting hyperinsulinemia in the presence of slight hypoglycemia (Fig. 2A and E), again showing increased insulin secretion. Plasma glucagon levels were unchanged in the fasted and fed condition (Fig. 2B) suggesting that α-cell function is not affected in Mitfce/ce animals. After intraperitoneal glucose challenge, glucose tolerance was improved (Fig. 2C and D), which also reflects increased glucose-stimulated insulin secretion. However, when glucose-stimulated insulin secretion was determined the only significant difference was observed at 0 min (Fig. 2E). Mitf function in peripheral tissues may also influence glucose responsiveness and partially account for the altered glucose clearance in Mitfce/ce. However, thus far, no data supporting a role of Mitf in muscle, liver, and adipose glucose metabolism have been reported, and transcription in these tissues is relatively low (gene expression atlas, http://www.ebi.ac.uk/arrayexpress/). Collectively, these observations suggest that loss of Mitf specifically alters β-cell function by increasing glucose responsiveness and insulin secretion.
Gene expression analysis showed increased transcription of key β-cell genes such as Pax4, Pax6, insulin, and Glut2, which could contribute to the increased insulin secretory response. Thus elevated insulin mRNA levels may result in increased total insulin content and in a larger pool of insulin granules that are ready to be released, which could influence insulin secretion. However, Mitfce/ce β-cells have no change in the total number of insulin granules (Supplementary Fig. 4 and Supplementary Table 1), and the percentage of ready releasable granules stays the same suggesting that mutant β-cell function is most likely improved owing to changes in glucose sensing. Thus, elevated Glut2 expression may positively influence this process, since it is the only glucose transporter expressed in β-cells that mediates glucose-stimulated insulin secretion (29,30).
Upregulation of Pax6 transcription may be the key to the improved β-cell function observed in Mitfce/ce, since it has been shown that this transcription factor regulates the expression of several β-cell genes like insulin, Glut2, PC1/3, and glucokinase (31). Interestingly, we observed enhanced transcriptional activity of only some Pax6 target genes like insulin and Glut2 but not others, which suggests that a twofold increase in Pax6 expression is not sufficient to augment the transcription of all Pax6 target genes in this context.
Loss-of-function studies have established a critical role for Pax4 in β-cell development (32), while ectopic Pax4 expression in adult β-cells is associated with increased proliferation and protection against apoptosis (33). Mitf binds to Pax4 regulatory elements, and Mitfce/ce islets have twofold higher Pax4 mRNA levels than wild-type islets but no difference in β-cell mass and thus no difference in β-cell proliferation and specification. This may be due to the moderate upregulation of Pax4 transcription in Mitfce/ce islets compared with Pax4 overexpression models used in recent work, which has studied the effects of 15-fold increased Pax4 (33). However, the moderately increased Pax4 expression in Mitfce/ce islets may still positively influence β-cell function and contribute to improved glucose clearance.
Previous studies have demonstrated that Mitf acts both as transcriptional activator and repressor (14,15,34): it activates genes required for cell pigmentation (13,14), while ectopic Mitf expression prevents Pax6 transcription in the developing eye (14,23) and Pax6 and Mitf cooperatively repress common target genes in the retinal pigmented epithelium (15). Thus far, Mitf repressor function has only been described in the context of coexpression with Pax6 but not with other transcriptional repressors like groucho (15,35). Here, we demonstrate that Mitf binds to and represses the Pax6 P0 regulatory region, which is required for high Pax6 expression during pancreas development (25,36). This finding suggests that Mitf is partially responsible for inactivation of the embryonic pancreatic Pax6 P0 promoter.
In vitro transfection and gel shift experiments have demonstrated that direct protein-protein interaction between Pax6 and Mitf results in an inactivation of both proteins (37). The Mitfce protein lacks the leucine zipper domain, which results in loss of DNA binding and reduced interaction with cofactors (19). The leucine zipper domain is also required for interactions with Pax6 (37); thus, Mitfce protein not only has lost its transcriptional activity but also fails to interact and repress Pax6 function in mutant β-cells. These in vitro studies support our observation that the expression of the newly identified direct Mitf target gene Pax6 is elevated in Mitfce/ce animals and that enhanced Pax6 levels and transcriptional activity may result in increased Glut2, Pax4, and insulin transcription.
Our data identify Mitf as a novel transcriptional repressor in adult β-cells and suggest that regulation of β-cell function depends on interplay between the Mitf and Pax6 transcription factors.
ACKNOWLEDGMENTS
This work was supported by the Swedish Research Council, the JDRF, Diabetesfonden, Lindhés, and Jeanssons Stiftelse.
No potential conflicts of interest relevant to this article were reported.
M.A.M. designed research, researched data, and wrote the manuscript. M.W., E.G., and J.K.C. researched data. J.K.J. reviewed the manuscript and contributed to discussion. H.B. researched data. M.F. researched data, reviewed the manuscript, and contributed to discussion. U.A.N. reviewed the manuscript and contributed to discussion. I.A. designed research and wrote the manuscript. M.A.M. and I.A. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank S. Saule (Institut Curie), C. Wright (Department of Cell and Developmental Biology, Vanderbilt University Medical Center), and the Beta Cell Biology Consortium (BCBC) for providing antibodies; B. Ahrén (Department of Clinical Sciences, Lund University), H. Semb (Stem Cell Center, Lund University, and The Danish Stem Cell Center, Copenhagen University), and R. Stein (Department of Molecular Physiology and Biophysics, Vanderbilt University School of Medicine) for comments on the manuscript; M. Magnusson for mouse strain maintenance; B.-M. Nilsson (Clinical Research Center, Lund University) for plasma glucagon measurements; and R. Wallén (Lund University) for transmission electron microscopy analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-1464/-/DC1.
REFERENCES
- 1.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994;371:606–609 [DOI] [PubMed] [Google Scholar]
- 2.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev 1998;12:1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashery-Padan R, Zhou X, Marquardt T, et al. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol 2004;269:479–488 [DOI] [PubMed] [Google Scholar]
- 4.Gu C, Stein GH, Pan N, et al. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab 2010;11:298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet 1997;17:138–139 [DOI] [PubMed] [Google Scholar]
- 6.Yasuda T, Kajimoto Y, Fujitani Y, et al. PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes 2002;51:224–230 [DOI] [PubMed] [Google Scholar]
- 7.Malecki MT, Jhala US, Antonellis A, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet 1999;23:323–328 [DOI] [PubMed] [Google Scholar]
- 8.Hole RL, Pian-Smith MC, Sharp GW. Development of the biphasic response to glucose in fetal and neonatal rat pancreas. Am J Physiol 1988;254:E167–E174 [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka T, Zhao L, Artner I, et al. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. 2003;23:6049–6062 [DOI] [PMC free article] [PubMed]
- 10.Matsuoka TA, Kaneto H, Stein R, et al. MafA regulates expression of genes important to islet beta-cell function. Mol Endocrinol 2007;21:2764–2774 [DOI] [PubMed] [Google Scholar]
- 11.Artner I, Blanchi B, Raum JC, et al. MafB is required for islet beta cell maturation. Proc Natl Acad Sci USA 2007;104:3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artner I, Hang Y, Mazur M, et al. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes 2010;59:2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibahara S, Yasumoto K, Amae S, et al. Regulation of pigment cell-specific gene expression by MITF. Pigment Cell Res 2000;13(Suppl. 8):98–102 [DOI] [PubMed] [Google Scholar]
- 14.Spence JR, Madhavan M, Aycinena J-C, Del Rio-Tsonis K. Retina regeneration in the chick embryo is not induced by spontaneous Mitf downregulation but requires FGF/FGFR/MEK/Erk dependent upregulation of Pax6. Mol Vis 2007;13:57–65 [PMC free article] [PubMed] [Google Scholar]
- 15.Bharti K, Gasper M, Ou J, et al. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet 2012;8:e1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 2002;109:707–718 [DOI] [PubMed] [Google Scholar]
- 17.Du J, Widlund HR, Horstmann MA, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 2004;6:565–576 [DOI] [PubMed] [Google Scholar]
- 18.Loercher AE, Tank EMH, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol 2005;168:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemesath TJ, Steingrímsson E, McGill G, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994;8:2770–2780 [DOI] [PubMed] [Google Scholar]
- 20.Johansson JK, Voss U, Kesavan G, et al. N-cadherin is dispensable for pancreas development but required for beta-cell granule turnover. Genesis 2010;48:374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahlenius H, Devaraju K, Monni E, et al. Adaptor protein LNK is a negative regulator of brain neural stem cell proliferation after stroke. J Neurosci 2012;32:5151–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnes L, Hill JT, Gross S, Magnuson MA, Sussel L. Ghrelin expression in the mouse pancreas defines a unique multipotent progenitor population. PLoS ONE 2012;7:e52026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mochii M, Mazaki Y, Mizuno N, Hayashi H, Eguchi G. Role of Mitf in differentiation and transdifferentiation of chicken pigmented epithelial cell. Dev Biol 1998;193:47–62 [DOI] [PubMed] [Google Scholar]
- 24.Morgan R. Conservation of sequence and function in the Pax6 regulatory elements. Trends Genet 2004;20:283–287 [DOI] [PubMed] [Google Scholar]
- 25.Carbe C, Hertzler-Schaefer K, Zhang X. The functional role of the Meis/Prep-binding elements in Pax6 locus during pancreas and eye development. Dev Biol 2012;363:320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steingrímsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 2004;38:365–411 [DOI] [PubMed] [Google Scholar]
- 27.White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH. Defining pancreatic endocrine precursors and their descendants. Diabetes 2008;57:654–668 [DOI] [PubMed] [Google Scholar]
- 28.Johnson SL, Nguyen AN, Lister JA. mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev Biol 2011;350:405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orci L, Thorens B, Ravazzola M, Lodish HF. Localization of the pancreatic beta cell glucose transporter to specific plasma membrane domains. Science 1989;245:295–297 [DOI] [PubMed] [Google Scholar]
- 30.Guillam MT, Dupraz P, Thorens B. Glucose uptake, utilization, and signaling in GLUT2-null islets. Diabetes 2000;49:1485–1491 [DOI] [PubMed] [Google Scholar]
- 31.Gosmain Y, Katz LS, Masson MH, Cheyssac C, Poisson C, Philippe J. Pax6 is crucial for β-cell function, insulin biosynthesis, and glucose-induced insulin secretion. Mol Endocrinol 2012;26:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature 1997;386:399–402 [DOI] [PubMed] [Google Scholar]
- 33.Hu He KH, Lorenzo PI, Brun T, et al. In vivo conditional Pax4 overexpression in mature islet β-cells prevents stress-induced hyperglycemia in mice. Diabetes 2011;60:1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev 2000;14:1712–1728 [PubMed] [Google Scholar]
- 35.Planque N, Raposo G, Leconte L, Anezo O, Martin P, Saule S. Microphthalmia transcription factor induces both retinal pigmented epithelium and neural crest melanocytes from neuroretina cells. J Biol Chem 2004;279:41911–41917 [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev 2002;16:2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planque N, Leconte L, Coquelle FM, Martin P, Saule S. Specific Pax-6/microphthalmia transcription factor interactions involve their DNA-binding domains and inhibit transcriptional properties of both proteins. J Biol Chem 2001;276:29330–29337 [DOI] [PubMed] [Google Scholar]