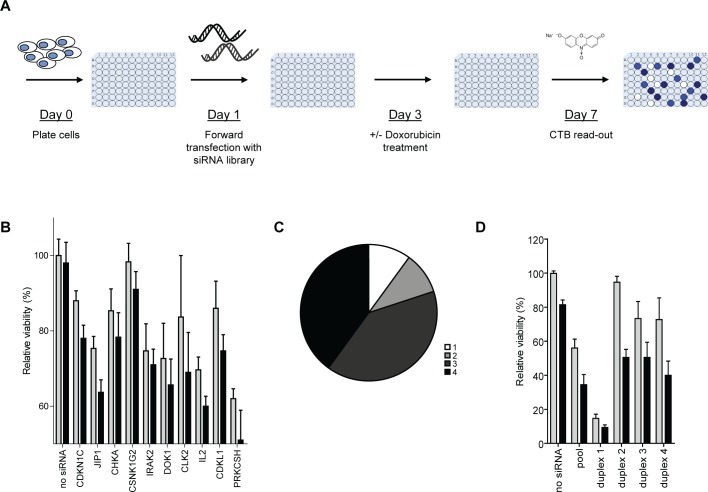
Figure 1. siRNA library screen of the human kinome identifies enhancers of doxorubicin response in OS.
(A) Schematic overview of the set-up of the screens performed with pools of 4 siRNAs against 788 human kinases and kinase-associated genes in SaOS-2 cells. (B) Screen results of the 10 selected candidate hits showing the effects of gene-silencing only (grey bars) versus gene-silencing + doxorubicin treatment (black bars) on cell viability. Bars represent the average cell viability measured in the 3 screens; error bars indicate standard deviations (SD). (C) Pie chart summarizing the secondary screen results. The chart shows the number of separate siRNA duplexes of 4 individual siRNAs tested per gene, that reproduced the doxorubicin-sensitising phenotype of the pooled siRNAs per gene. (Bar graphs for the separate siRNA duplexes are provided in panel D for JIP1 and Supplementary Figure S1 for the other genes). (D) Confirmation of the doxorubicin-sensitising phenotype with siRNAs against JIP1. Cells were transfected with the indicated siRNA duplex and cultured in the presence (black bars) or absence (grey bars) of doxorubicin at IC20 concentration. Bars represent results from an experiment performed in triplicate; error bars indicate SD..