Fig. 3.
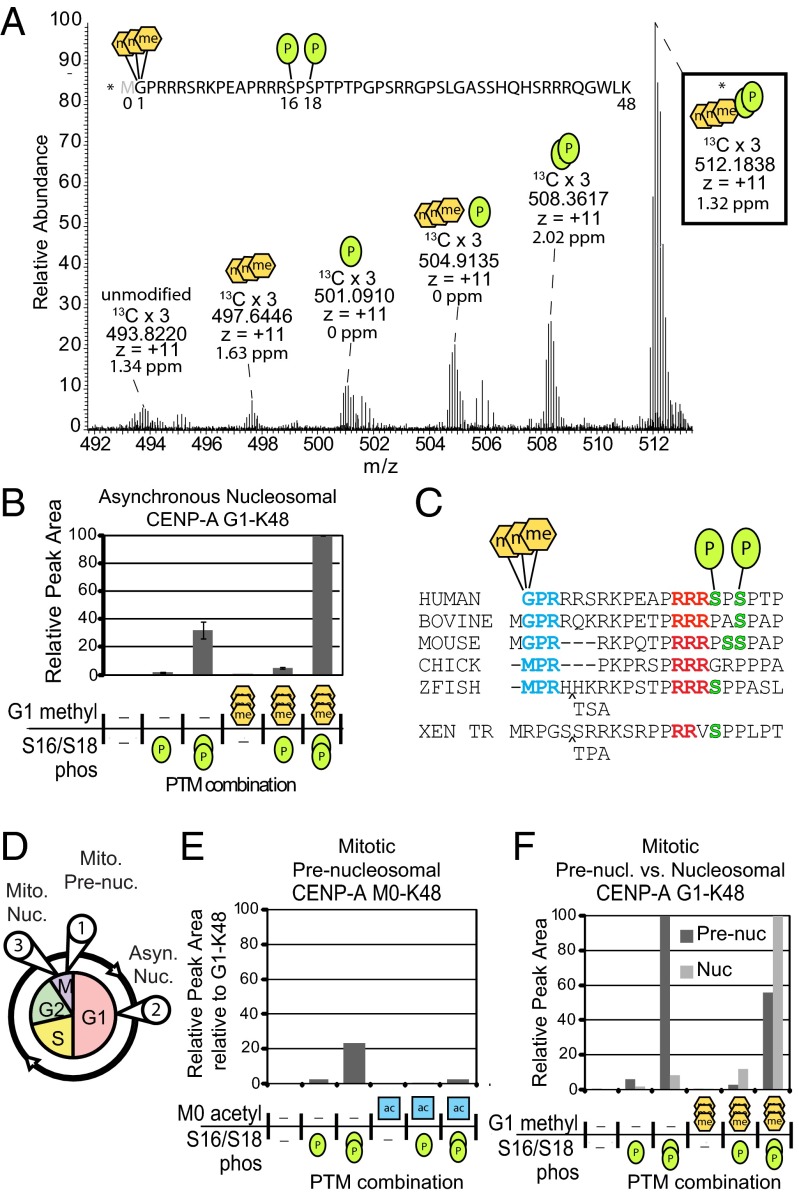
CENP-A N-terminal tails are marked with combinations of stable phosphorylation and α-N methylation. (A) LysC digestion yields a CENP-A G1–K48 peptide. A full MS spectrum (17 averaged scans) shows combinatorial α-N trimethylation and phosphorylations of the same peptides in asynchronously cycling cells. Accurate mass and charge state is reported for the 13C x 3 isotopes. The most abundant form is trimethylated at Gly1 and doubly phosphorylated at Ser16 and Ser18. Green oval, phosphorylation; orange hexagon, methylation. (B) Integrated chromatographic peak areas of CENP-A G1–K48 PTM forms. n = 3 independent biological replicates. Error bars represent SEM. (C) Comparison of vertebrate CENP-A protein sequences reveals conservation at observed PTM sites: N termini (blue), proline–arginine (red), and serine (green). (D) Soluble and chromatin fractions of cells blocked in mitosis were used to purify populations of (circle 1) prenucleosomal, and nucleosomal CENP-A from (circle 2) asynchronously-dividing cells and (circle 3) mitotic cells. Integrated chromatographic peak area quantified CENP-A tail PTM forms: (E) prenucleosomal containing Metinit; (F) prenucleosomal and mitotic nucleosomal fractions lacking Metinit. Acetylation (blue square) was observed only when Metinit was present.