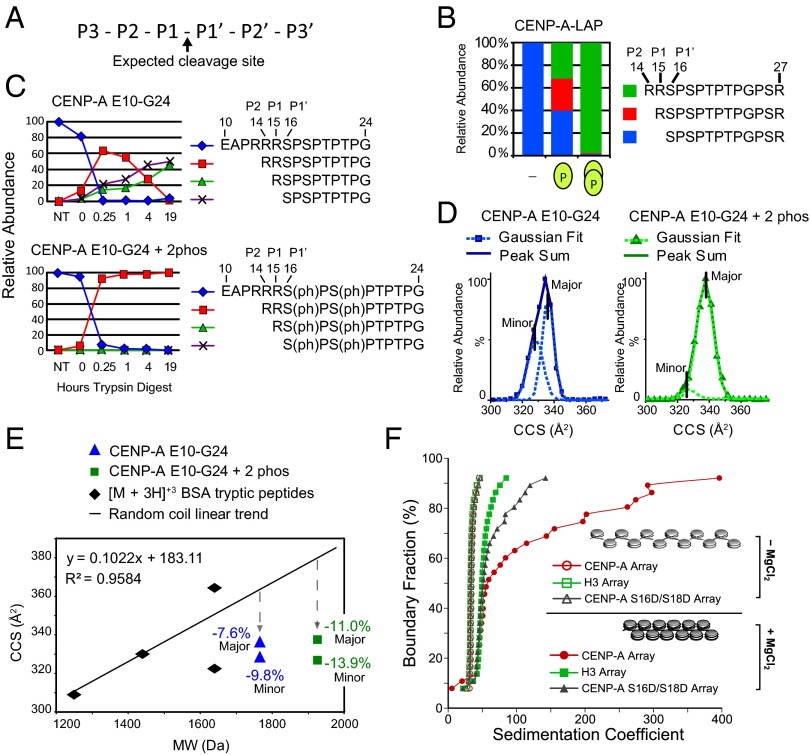
Fig. 4.
CENP-A Ser16/Ser18 phosphorylation forms a compact secondary structure. (A) Nomenclature for proteolytic sites. (B) The percentage of trypsin cleavage at the P1–P1′, P1–P2, and P2-P3 sites for peptides derived from affinity-purified CENP-A. (C) The percentage of trypsin cleavage at the P1–P1′, P1–P2, and P2–P3 sites for unmodified and phosphorylated synthetic CENP-A peptides E10–G24 digested with trypsin over time were followed by liquid chromatography (LC)-MS. (D) IM-MS analysis of the monomeric [M + 3H]+3 ion of unmodified and doubly phosphorylated E10–G24 peptide is used to calculate the ion-neutral collision cross section (CCS) for each species. CCS values for the major- and minor-contributing conformers were calculated as Gaussian peaks from the sum of each peptide. (E) Tryptic digested BSA peptides analyzed by IM-MS are plotted according to CCS and molecular weight. Comparison of phosphorylated (square) and unmodified (triangle) E10–G24 peptide with the random coil trend line reveals that phosphorylation causes greater compactness relative to the increase in molecular weight. (F) AUC profiles of in vitro assembled chromatin arrays showing differential MgCl2-dependent folding between H3, CENP-A, and phospho-mimetic (S16D/S18D) CENP-A arrays. All array types (minus MgCl2) have overlapping sedimentation profiles, indicating that they are saturated to the same extent. Therefore, the observed differences upon array folding are exclusively due to the contributions of different proteins.