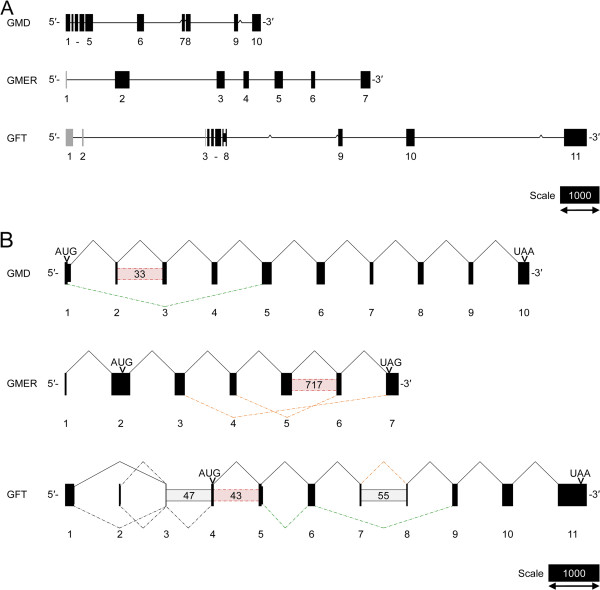
Figure 2.
Genomic organization and splicing of GDP-L-fucose synthesis- and transport-associated genes in Schistosoma mansoni. The mRNA transcript sequences of GMD, GMER and GFT were mapped onto genomic scaffolds in the SchistoDB [35](A). Exons (boxes, numbered below) and introns (connecting lines) are drawn to scale (bar = 1000 nt) with GMD/GMER/GFT-coding elements, including exons and a subset of retained introns, depicted as black boxes and non-coding exons depicted as gray boxes. Caret marks indicate gaps in the genomic sequence. Alternative splicing, including exon skipping, intron retention, mutual exclusivity and use of alternate splice donor sites, was observed during transcript sequencing (B). Bent connectors indicate splicing between exons (boxes, numbered below), with solid lines representing splicing in the main/major full-length GMD/GMER/GFT-coding transcripts and dotted lines representing alternative splice events. In this panel, exons are drawn to scale (bar = 1000 nt) and spacing of exons is arbitrary. Interexonic boxes represent retained introns (estimated lengths in parentheses) with solid outlines signifying retention in the main/major transcript and dotted lines indicating retention in other isoforms. The positions of the prototypical start and stop codons (AUG and UAA/UAG, respectively) are shown. Colors convey the in silico consequences of splicing: black, conservation of the prototypical ORF; red, introduction of a PTC; orange, induction of a downstream frameshift; green, in-frame deletion/addition.