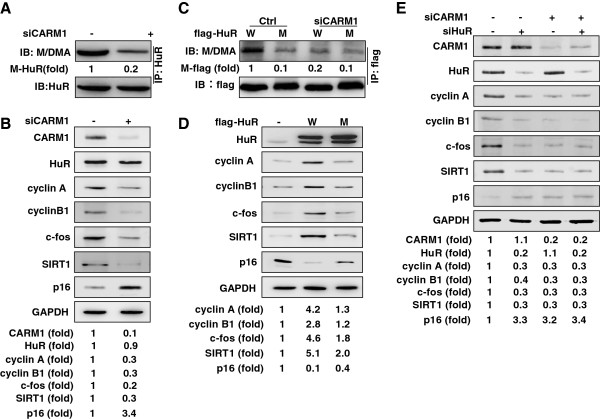
Figure 1.
Methylation by CARM1 enhances the regulating of cyclin A, cyclin B1, c-fos, SIRT1, and p16 by HuR. (A) Forty-eighth after transfection of HeLa cells with CARM1siRNA (+) or a control siRNA (−), lysates were prepared for IP assays by using HuR antibody. The presence of total and methylated HuR in the IP materials was determined by Western blot analysis by using M/DMA and HuR antibodies, respectively. (B) Cell lysates described in Figure 1A were subjected to Western blot analysis to assess the protein levels of CARM1, HuR, cyclin A, cyclin B1, c-fos, SIRT1, p16, and GAPDH. Western blotting signals were quantified by densitometry. (C) HeLa cells were transfected with a vector expressing flag-HuR or flag-HuRΔ. Twenty fourh later, cells were further transfected with CARM1 siRNA or a control siRNA and cultured for an additional 48 h. Whole-cell lysates were prepared and subjected to IP assays by using anti-flag antibody (M2). Western blot analysis was performed to assess the total and methylation levels of flag-tagged HuR in the IP materials using M/MDA and flag antibodies, respectively. (D) HeLa cells were transfected with a vector expressing flag-HuR or flag-HuRΔ. Forty eighth later, lysates were prepared to assess the protein levels of CARM1, HuR, cyclin A, cyclin B1, c-fos, SIRT1, p16, and GAPDH by Western blot analysis, the signals of Western blotting were quantified by densitometry. (E) HeLa cells were either transfected with HuR or CARM1 siRNA or co-transfected with both siRNAs. Forty eighth later, Western blot analysis was performed to evaluate the levels of CARM1, HuR, cyclin A, cyclin B1, c-fos, SIRT1, p16, and GAPDH; Western blotting signals were quantified by densitometry. Data are representatives from 3 independent experiments.