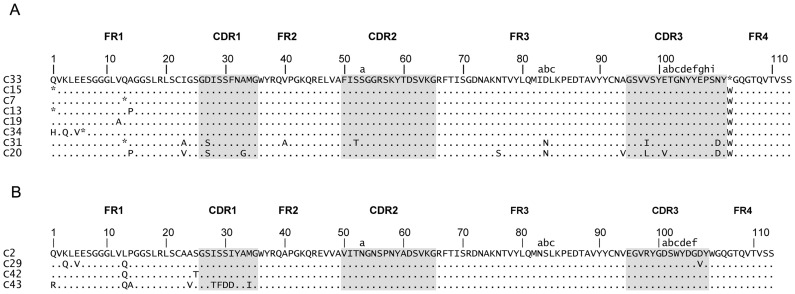
Figure 4. Amino acid sequence alignment of anti-α–Cbtx VHH binders.
The clones were categorized in either (A) Cluster I or (B) Cluster II based on their sequence homologies. Framework regions (FR) and complementarity determining regions (CDR) (shaded in grey) as well as amino acid insertions (52a, 82a, b, c, 100a, b, c, d, e, f, g, h, i) are defined according to the Kabat numbering system [52]. The dots in the sequences indicate amino acid identity that is the same as in C33 (Cluster I) or C2 (Cluster II). All clones belong to VHH Subfamily 2 [49]. An asterisk (*) represents an amber stop codon (TAG) mutation. However, TG1 E. coli, which was used during panning, has the supE amber suppressor mutation (now known as glnV gene for tRNAgln gene) that inserts a glutamine (Q) when stop codon readthrough occurs [53]. Therefore, a glutamine (Q) would have been placed were asterisks are marked, thus allowing these clones to survive the panning process.