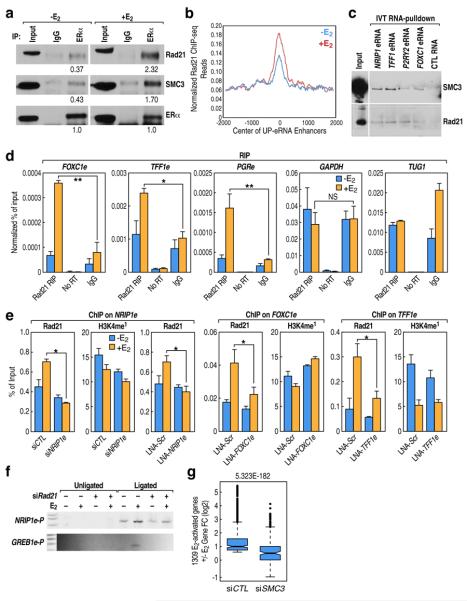
Fig4. Role of eRNA in Cohesin-dependent gene activation.
(a) Co-immunoprecipitation of Rad21 and SMC3 with ERα from E2 or ethanol treated MCF7 whole cell extracts showing physical interaction between ERα and Cohesin subunits, which is further enhanced by E2 treatment. Numbers below blots depict the band density (Image J) relative to that of corresponding density of ERα. (b) Rad21 enrichment centered at UP-enhancers as determined by ChIP-seq, which shows moderate E2-induced increase. (c) In vitro transcribed (IVT) RNA pull-down assay showing the interaction between Cohesin subunits and eRNAs, but not a control RNA (RNA fragment of Xenopus elongation factor α). (d) RIP-QPCR showing binding of Rad21 to selected regulated eRNAs but not to GAPDH or TUG1. (e) ChIP-QPCR analyses represent the inhibitory effect from knock-down of NRIP1e (siRNA and LNA), FOXC1e or TFF1e on E2-indcued Rad21 additional recruitment, but not on H3K4me1. (f) Effect of Rad21 depletion on physical interaction between promoter:enhancer for the GREB1 and NRIP1 genes, assessed by 3C assay. (g) 1,309 E2-induced coding genes were defined by GRO-seq from siCTL (±E2) group and then their fold change (log2 FC) in siCTL versus siSMC3 transfected MCF7 cells was plotted). Mean ± SD. (n=3). *p<0.05, **p<0.01.