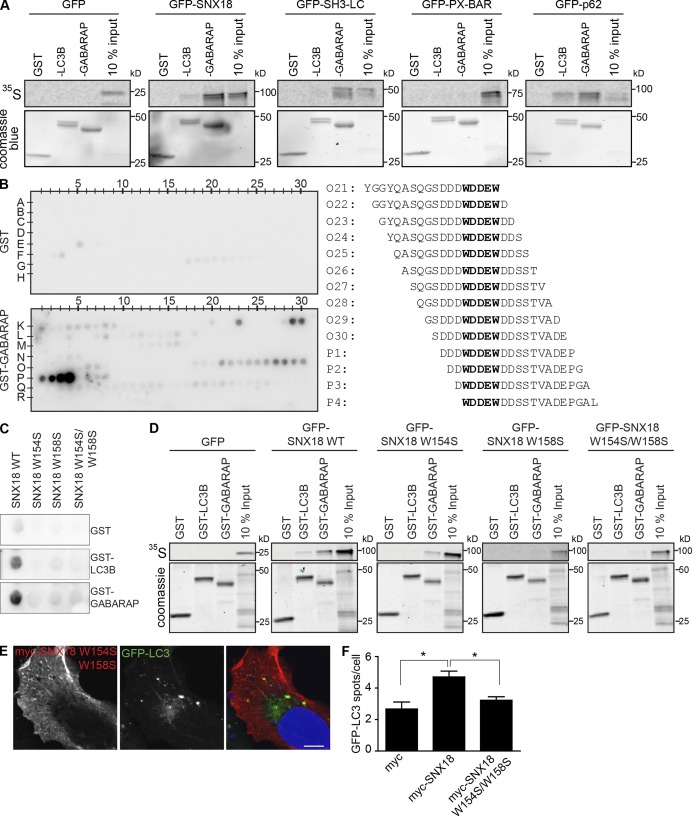
Figure 7.
A WDDEW sequence in the SNX18 LC region mediates the interaction with LC3/GABARAP. (A) GFP-p62 or -SNX18 full-length, SH3-LC, or PX-BAR regions were in vitro translated and incubated with GST-LC3B or -GABARAP. The resulting pulldowns were separated by SDS-PAGE. Bound proteins were detected by autoradiography and GST proteins by Coomassie blue staining. (B) 18-mer peptides covering the entire sequence of the SNX18 SH3-LC region were spotted on a membrane that was incubated with GST or GST-GABARAP, which were detected by immunoblotting against GST. The peptide sequences that specifically bound GST-GABARAP are shown with the common WDDEW motif in bold. (C) Peptides with the sequence YGGYQASQGSDDDWDDEWDDSSTVADEPGAL (SNX18 WT) or with the first (SNX18 W154S), the second (SNX18 W158S), or both (SNX18 W154S/W158S) W mutated to S were spotted on membranes that were incubated with GST, GST-LC3B, or GST-GABARAP. Binding was analyzed as in B. (D) GFP-SNX18 WT and the indicated mutants were in vitro translated and incubated with GST-LC3B or -GABARAP, and their binding was analyzed as in A. (E) HeLa GFP-LC3 cells were transfected with myc-SNX18 W154S/W158S mutant, starved for 2 h, immunostained against myc, and analyzed by confocal imaging. Bar, 10 µm. (F) Indicated myc-SNX18 constructs were transfected into HEK GFP-LC3 cells, and the number of GFP-LC3 spots per cell was quantified. The graph shows mean ± SEM (error bars), n = 3. *, P < 0.05.