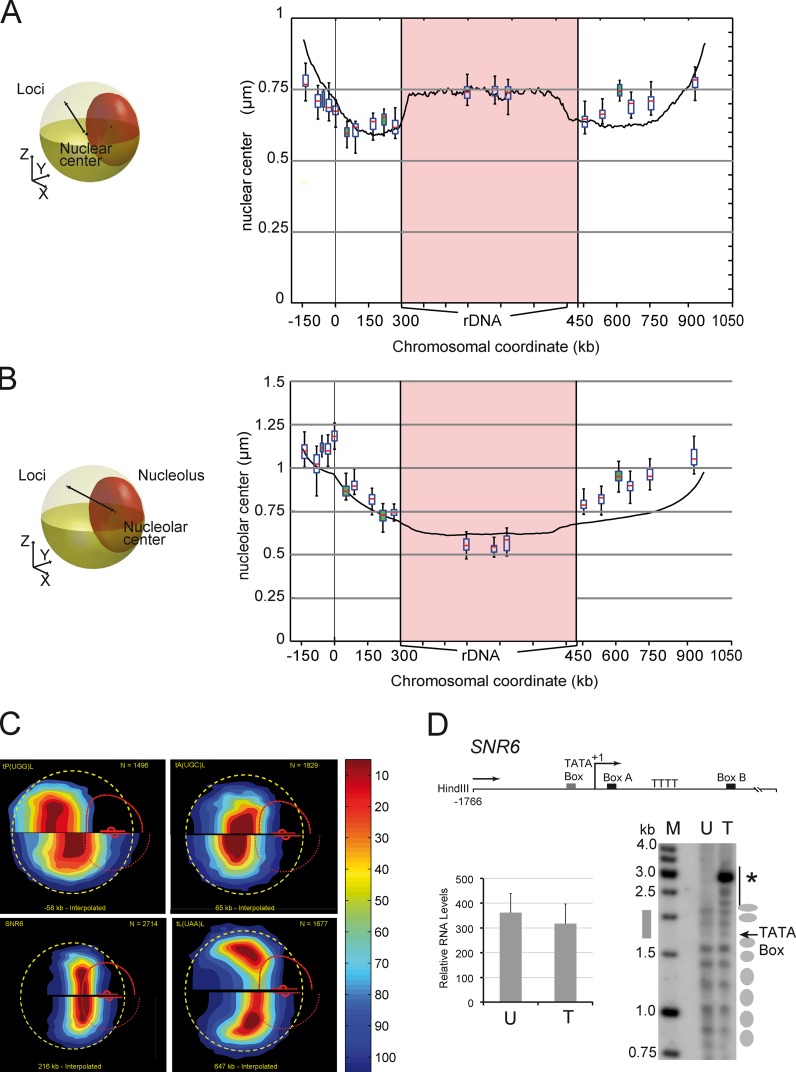
Figure 2.
The position of loci along chromosome XII relative to the NUC and the NE are predicted by computational modeling. (A and B) The distance of the locus to the nuclear center (A) and to the nucleolar center (B) is plotted versus its genomic position. Yellow and red ellipsoids depict the NE and NUC, respectively, and the black line represents the distance (left). The median distance is shown with box plots for the 15 loci described in Fig. 1 and for four RNA Pol III genes (white and green datasets). The median distance of chromosome XII loci to the nuclear center and to the centroid of the rDNA segment from a computational model of chromosome XII (Wong et al., 2012) is shown with solid black lines. (C) The gene map of loci t(P(UGG)L, tA(UGC)L, SNR6, and t(L(UAA)L relative to interpolated positions −58, 65, 216, and 647 kb (which correspond to their genomic coordinates) reveals global agreement between the model and measurements (see Video 2). The experimentally determined gene map (top) is compared with the interpolated position (bottom). The dashed yellow circle, the curved broken red line, and the small red dot depict the median NE, the median NUC, and the median location of the nucleolar center, respectively. (D) The yeast U6 small nuclear RNA (snRNA) gene SNR6 is unaffected by FROS insertion. Locations of important elements are indicated relative to the transcriptional start site (bent arrow) as +1 (top). Indirect end-labeling analysis of the chromatin structure on SNR6 is shown on the bottom-right panel. Ellipses on the right mark the positioned nucleosomes in the corresponding lanes with wild-type unmodified (U) or FROS tagged (T) strains. Note that the apparent hyper-sensitive site (asterisk) detected upon FROS insertion corresponds to a HindIII site introduced along with tetO repeat. SNR6 expression is not affected by FROS insertion. Transcript levels in untagged and FROS-tagged strains were normalized against the U4 transcript used as an internal control. The mean and scatter of RNA levels estimated from three independent experiments are plotted (error bars; bottom left).