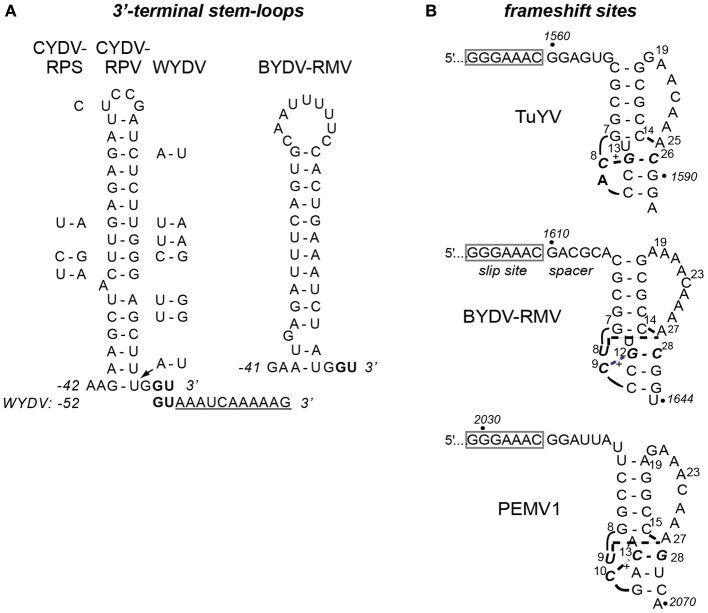
Figure 6.
Predicted secondary structures in BYDV-RMV RNA. (A) Stem-loop at the 3′ terminus of CYDV-RPV genome as determined by Osman et al. (2006), flanked by base differences in CYDV-RPS (left) and WYDV (right) which show covariations in base pairing that maintain secondary structure. Conserved 3′ terminal bases, GU are shown in bold. Eleven extra bases at the 3′ end of the WYDV genome, not present in any other polerovirus, are shown below the CYDV-RPV sequence in underlined text. The proposed secondary structure of the 3′ end of the BYDV-RMV genome was predicted using Mfold (Zuker, 2003). Base numbering (negative) is from the 3′ end of the genomes. (B) Predicted (BYDV-RMV) and known (PEMV1 and TuYV) (Su et al., 1999; Nixon et al., 2002; Miller and Giedroc, 2010) tertiary structures of pseudoknots downstream of the frameshift sites (boxed). Italics indicate base numbers in the genome. Other numbering is the position in the fragment used for nmr (except BYDV-RMV where the number allows comparison with the other structures). Short curved lines indicate phosphodiester backbone as necessary for two-dimensional rendering. Bold, dashed lines indicate non-Watson-Crick interactions between bases. +indicated protonated cytidine that participates in base triples. Due to the recent change of the name of the BWYV isolate used in previous structural studies (Domier, 2012) it is now indicated by the new name, TuYV.